[English] 日本語
 Yorodumi
Yorodumi- PDB-2gvo: Solution structure of a purine rich hexaloop hairpin belonging to... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2gvo | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides | ||||||
 Components Components | GUANOSINE-5'-MONOPHOSPHATE | ||||||
 Keywords Keywords | RNA / RNA structure / hairpin structure / G.U wobble pair / hexaloop / 13C/15N-labeled RNA | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
 Authors Authors | Joli, F. / Bouchemal, N. / Laigle, A. / Hartmann, B. / Hantz, E. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2006 Journal: Nucleic Acids Res. / Year: 2006Title: Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides. Authors: Joli, F. / Bouchemal, N. / Laigle, A. / Hartmann, B. / Hantz, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2gvo.cif.gz 2gvo.cif.gz | 267.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2gvo.ent.gz pdb2gvo.ent.gz | 200.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2gvo.json.gz 2gvo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gv/2gvo https://data.pdbj.org/pub/pdb/validation_reports/gv/2gvo ftp://data.pdbj.org/pub/pdb/validation_reports/gv/2gvo ftp://data.pdbj.org/pub/pdb/validation_reports/gv/2gvo | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
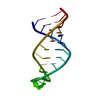
| Deposited unit | 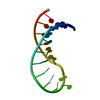
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 5844.515 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: This sequence occurs naturally in Homo sapiens (humans) |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: The structure was determined using triple-resonance NMR spectroscopy |
- Sample preparation
Sample preparation
| Details | Contents: 1mM 18mer mRNA uniformly 13C/15N labeled, 10 mM Sodium phosphate buffer, 90% H2O, 10% D2O Solvent system: 90% H2O/10% D2O | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 Details: the structures are based on a total of 187 restraints, 84 are sequential and long range internucleotide distances, 85 dihedral angle restraints, 18 sugar puckers restraints and 15 distance ...Details: the structures are based on a total of 187 restraints, 84 are sequential and long range internucleotide distances, 85 dihedral angle restraints, 18 sugar puckers restraints and 15 distance restaints from hydrogen bonds. | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted, back calculated data agree with experimental NOESY spectrum, structures with acceptable covalent geometry, structures with favorable ...Conformer selection criteria: all calculated structures submitted, back calculated data agree with experimental NOESY spectrum, structures with acceptable covalent geometry, structures with favorable non-bond energy,structures with the least restraint violations, structures with the lowest energy Conformers calculated total number: 30 / Conformers submitted total number: 21 |
 Movie
Movie Controller
Controller






 PDBj
PDBj





























 HSQC
HSQC