+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2gd3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of S14G-humanin in 30% TFE solution | ||||||
 Components Components | Humanin | ||||||
 Keywords Keywords | UNKNOWN FUNCTION / S14G-Humanin / Humanin / Alzheimer's disease / Neuroprotection / NMR / CD | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of interleukin-18 production / receptor antagonist activity / negative regulation of neuroinflammatory response / negative regulation of interleukin-1 production / negative regulation of NLRP3 inflammasome complex assembly / negative regulation of execution phase of apoptosis / negative regulation of amyloid fibril formation / Formyl peptide receptors bind formyl peptides and many other ligands / leukocyte chemotaxis / negative regulation of response to oxidative stress ...negative regulation of interleukin-18 production / receptor antagonist activity / negative regulation of neuroinflammatory response / negative regulation of interleukin-1 production / negative regulation of NLRP3 inflammasome complex assembly / negative regulation of execution phase of apoptosis / negative regulation of amyloid fibril formation / Formyl peptide receptors bind formyl peptides and many other ligands / leukocyte chemotaxis / negative regulation of response to oxidative stress / sperm flagellum / supramolecular fiber organization / sperm midpiece / mitochondrion organization / G protein-coupled receptor binding / negative regulation of inflammatory response / cellular response to amyloid-beta / cell-cell signaling / G alpha (i) signalling events / G alpha (q) signalling events / negative regulation of neuron apoptotic process / intracellular iron ion homeostasis / signaling receptor binding / apoptotic process / negative regulation of apoptotic process / perinuclear region of cytoplasm / mitochondrion / extracellular space / extracellular region / identical protein binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Method | SOLUTION NMR / simulated annealing torsion angle dynamics | ||||||
 Authors Authors | Benaki, D. / Zikos, C. / Evangelou, A. / Livaniou, E. / Vlassi, M. / Mikros, E. / Pelecanou, M. | ||||||
 Citation Citation |  Journal: Biochem.Biophys.Res.Commun. / Year: 2006 Journal: Biochem.Biophys.Res.Commun. / Year: 2006Title: Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease. Authors: Benaki, D. / Zikos, C. / Evangelou, A. / Livaniou, E. / Vlassi, M. / Mikros, E. / Pelecanou, M. #1:  Journal: Biochem.Biophys.Res.Commun. / Year: 2005 Journal: Biochem.Biophys.Res.Commun. / Year: 2005Title: Solution structure of humanin, a peptide against Alzheimer's disease-related neurotoxicity. Authors: Benaki, D. / Zikos, C. / Evangelou, A. / Livaniou, E. / Vlassi, M. / Mikros, E. / Pelecanou, M. #2: Journal: Proc.Natl.Acad.Sci.Usa / Year: 2001 Title: A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Authors: Hashimoto, Y. / Niikura, T. / Tajima, H. / Yasukawa, T. / Sudo, H. / Ito, Y. / Kita, Y. / Kawasumi, M. / Kouyama, K. / Doyu, M. / Sobue, G. / Koide, T. / Tsuji, S. / Lang, J. / Kurokawa, K. / Nishimoto, I. #3:  Journal: Biochem.Biophys.Res.Commun. / Year: 2001 Journal: Biochem.Biophys.Res.Commun. / Year: 2001Title: Mechanisms of Neuroprotection by a Novel Rescue Factor Humanin from Swedish Mutant Amyloid Precursor Protein Authors: Hashimoto, Y. / Ito, Y. / Niikura, T. / Shao, Z. / Hata, M. / Oyama, F. / Nishimoto, I. | ||||||
| History |
|
- Structure visualization
Structure visualization
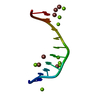
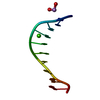
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2gd3.cif.gz 2gd3.cif.gz | 112.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2gd3.ent.gz pdb2gd3.ent.gz | 75.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2gd3.json.gz 2gd3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gd/2gd3 https://data.pdbj.org/pub/pdb/validation_reports/gd/2gd3 ftp://data.pdbj.org/pub/pdb/validation_reports/gd/2gd3 ftp://data.pdbj.org/pub/pdb/validation_reports/gd/2gd3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2661.239 Da / Num. of mol.: 1 / Mutation: S14G / Source method: obtained synthetically Details: S14G-humanin was prepared by Fmoc-solid phase synthesis on o-Cl-trityl-amidomethyl polystyrene resin (Evangelou, A., Zikos, C., Livaniou, E., Evangelatos, G.P. "Highyield, solid-phase ...Details: S14G-humanin was prepared by Fmoc-solid phase synthesis on o-Cl-trityl-amidomethyl polystyrene resin (Evangelou, A., Zikos, C., Livaniou, E., Evangelatos, G.P. "Highyield, solid-phase synthesis of humanin, an Alzheimer's disease associated, novel 24-mer peptide which contains a difficult sequence" J. Peptide Sci. 2004, 10, 631 635). The peptide was purified to 95% with semi-preparative RP-HPLC and suitably characterized. The sequence is naturally found in Homo sapiens (human). References: UniProt: Q8IVG9 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques |
- Sample preparation
Sample preparation
| Details | Contents: 1.3 mM S14G-humanin; 60 microM NaN3 to prevent microbial growth; H2O/TFE-d3 7:3; pH 2.6 (uncorrected for the presence of TFE); at 298 and 280 K Solvent system: H2O/TFE-d3 7:3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing torsion angle dynamics / Software ordinal: 1 Details: Starting from an extended structure a total of 300 structures were generated from 133 NOE-derived distance restraints and 2 distance restraints based on temperature coefficient data using ...Details: Starting from an extended structure a total of 300 structures were generated from 133 NOE-derived distance restraints and 2 distance restraints based on temperature coefficient data using the simulated annealing and energy minimization protocol in the program CNS, version 1.1 | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: 14 convergent conformers are presented having the lowest energy and the best structural quality in Ramachadran plot Conformers calculated total number: 300 / Conformers submitted total number: 14 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

