[English] 日本語
 Yorodumi
Yorodumi- PDB-2f77: Solution structure of the R55F mutant of M-PMV matrix protein (p10) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2f77 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of the R55F mutant of M-PMV matrix protein (p10) | ||||||
 Components Components | Core protein p10 | ||||||
 Keywords Keywords | VIRAL PROTEIN / 4 alpha-helices | ||||||
| Function / homology |  Function and homology information Function and homology informationviral budding via host ESCRT complex / viral nucleocapsid / structural constituent of virion / nucleic acid binding / host cell cytoplasm / viral translational frameshifting / zinc ion binding / metal ion binding Similarity search - Function | ||||||
| Biological species |  Simian retrovirus 1 Simian retrovirus 1 | ||||||
| Method | SOLUTION NMR / simulated annealing; molecular dynamics; torsion angle dynamics | ||||||
 Authors Authors | Vlach, J. / Lipov, J. / Veverka, V. / Lang, J. / Srb, P. / Rumlova, M. / Hunter, E. / Ruml, T. / Hrabal, R. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2008 Journal: Proc.Natl.Acad.Sci.USA / Year: 2008Title: D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein. Authors: Vlach, J. / Lipov, J. / Rumlova, M. / Veverka, V. / Lang, J. / Srb, P. / Knejzlik, Z. / Pichova, I. / Hunter, E. / Hrabal, R. / Ruml, T. | ||||||
| History |
|
- Structure visualization
Structure visualization
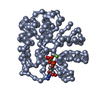
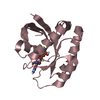
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2f77.cif.gz 2f77.cif.gz | 588.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2f77.ent.gz pdb2f77.ent.gz | 495.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2f77.json.gz 2f77.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f7/2f77 https://data.pdbj.org/pub/pdb/validation_reports/f7/2f77 ftp://data.pdbj.org/pub/pdb/validation_reports/f7/2f77 ftp://data.pdbj.org/pub/pdb/validation_reports/f7/2f77 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2f76C C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 11975.759 Da / Num. of mol.: 1 / Fragment: residues 1-100 of Gag polyprotein / Mutation: R55F Source method: isolated from a genetically manipulated source Details: A part of Core polyprotein which contains: Core protein p10; Core phosphoprotein pp18; Core protein p12; Core protein p27; Core protein p14; Core protein p4 Source: (gene. exp.)  Simian retrovirus 1 / Genus: Betaretrovirus / Species: Mason-Pfizer monkey virus / Gene: gag / Plasmid: pET22b / Species (production host): Escherichia coli / Production host: Simian retrovirus 1 / Genus: Betaretrovirus / Species: Mason-Pfizer monkey virus / Gene: gag / Plasmid: pET22b / Species (production host): Escherichia coli / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 1 mM matrix protein U-13C, U-15N; 100 mM phosphate buffer (K); 100 mM NaCl; 10 mM DTT; 95% H2O, 5% D2O Solvent system: 95% H2O/5% D2O |
|---|---|
| Sample conditions | Ionic strength: 600 mM / pH: 6 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing; molecular dynamics; torsion angle dynamics Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average, fewest violations, lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with acceptable covalent geometry, structures with favorable non-bond energy, structures with the least restraint violations, structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 18 |
 Movie
Movie Controller
Controller










 PDBj
PDBj

 NMRPipe
NMRPipe