[English] 日本語
 Yorodumi
Yorodumi- PDB-2cka: Solution structures of the BRK domains of the human Chromo Helica... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2cka | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | ||||||
 Components Components | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 8 | ||||||
 Keywords Keywords | HYDROLASE / BRK DOMAIN / TRANSCRIPTION ELONGATION / CHROMATIN REMODELING / PROTEIN-PROTEIN INTERACTION / TRANSCRIPTION / HELICASE / NUCLEAR PROTEIN / CHROMATIN REGULATOR | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of fibroblast apoptotic process / histone H3K4me3 reader activity / ATP-dependent chromatin remodeler activity / digestive tract development / positive regulation of transcription by RNA polymerase III / MLL1 complex / social behavior / prepulse inhibition / DNA helicase activity / Deactivation of the beta-catenin transactivating complex ...negative regulation of fibroblast apoptotic process / histone H3K4me3 reader activity / ATP-dependent chromatin remodeler activity / digestive tract development / positive regulation of transcription by RNA polymerase III / MLL1 complex / social behavior / prepulse inhibition / DNA helicase activity / Deactivation of the beta-catenin transactivating complex / negative regulation of canonical Wnt signaling pathway / beta-catenin binding / brain development / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Wnt signaling pathway / mRNA processing / p53 binding / histone binding / in utero embryonic development / chromatin remodeling / negative regulation of DNA-templated transcription / chromatin binding / positive regulation of DNA-templated transcription / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / protein-containing complex / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method | SOLUTION NMR / CANDID IN CYANA | ||||||
 Authors Authors | Ab, E. / de Jong, R.N. / Diercks, T. / Xiaoyun, J. / Daniels, M. / Kaptein, R. / Folkers, G.E. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction Authors: Ab, E. / De Jong, R.N. / Diercks, T. / Xiaoyun, J. / Daniels, M. / Kaptein, R. / Folkers, G.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
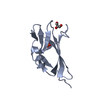
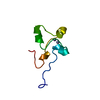
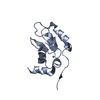
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2cka.cif.gz 2cka.cif.gz | 378 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2cka.ent.gz pdb2cka.ent.gz | 310.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2cka.json.gz 2cka.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ck/2cka https://data.pdbj.org/pub/pdb/validation_reports/ck/2cka ftp://data.pdbj.org/pub/pdb/validation_reports/ck/2cka ftp://data.pdbj.org/pub/pdb/validation_reports/ck/2cka | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 11059.638 Da / Num. of mol.: 1 / Fragment: BRK DOMAIN, RESIDUES 2012-2085 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) HOMO SAPIENS (human)Description: SELF MADE CDNA LIBRARY OF MULTIPLE HUMAN CARCINOMA CELL LINES Plasmid: PHISLIC / Production host:  References: UniProt: Q9HCK8, Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TRIPLE-RESONANCE NMR SPECTROSCOPY ON 13C, 15N-LABELED CHD8 2012-2085 |
- Sample preparation
Sample preparation
| Details | Contents: 95% H2O/5% D2O |
|---|---|
| Sample conditions | Ionic strength: 150 mM / pH: 7 / Pressure: 1.0 atm / Temperature: 298.0 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX / Manufacturer: Bruker / Model: DMX / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: CANDID IN CYANA / Software ordinal: 1 Details: EFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | |||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 50 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj