| Entry | Database: PDB / ID: 2c3b
|
|---|
| Title | The Crystal Structure of Aspergillus fumigatus Cyclophilin reveals 3D Domain Swapping of a Central Element |
|---|
 Components Components | PPIASE |
|---|
 Keywords Keywords | ISOMERASE / 3D DOMAIN SWAPPING / MISFOLDING / PPIASE / ASP F 11 / ALLERGEN / ROTAMASE |
|---|
| Function / homology |  Function and homology information Function and homology information
cyclosporin A binding / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / protein folding / mitochondrion / identical protein binding / cytoplasmSimilarity search - Function Cyclophilin-like / Cyclophilin / Cyclophilin-type peptidyl-prolyl cis-trans isomerase / Cyclophilin-type peptidyl-prolyl cis-trans isomerase, conserved site / Cyclophilin-type peptidyl-prolyl cis-trans isomerase signature. / Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain profile. / Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain / Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD / Cyclophilin-like domain superfamily / Beta Barrel / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |   ASPERGILLUS FUMIGATUS (mold) ASPERGILLUS FUMIGATUS (mold) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.85 Å MAD / Resolution: 1.85 Å |
|---|
 Authors Authors | Limacher, A. / Kloer, D.P. / Fluckiger, S. / Folkers, G. / Crameri, R. / Scapozza, L. |
|---|
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006
Title: The Crystal Structure of Aspergillus Fumigatus Cyclophilin Reveals 3D Domain Swapping of a Central Element
Authors: Limacher, A. / Kloer, D.P. / Fluckiger, S. / Folkers, G. / Crameri, R. / Scapozza, L. |
|---|
| History | | Deposition | Oct 5, 2005 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Jan 30, 2006 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 13, 2011 | Group: Advisory / Version format compliance |
|---|
| Revision 1.2 | Nov 13, 2024 | Group: Data collection / Database references ...Data collection / Database references / Other / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_entry_details / pdbx_modification_feature
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf |
|---|
|
|---|
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. |
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 1.85 Å
MAD / Resolution: 1.85 Å  Authors
Authors Citation
Citation Journal: Structure / Year: 2006
Journal: Structure / Year: 2006 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2c3b.cif.gz
2c3b.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2c3b.ent.gz
pdb2c3b.ent.gz PDB format
PDB format 2c3b.json.gz
2c3b.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/c3/2c3b
https://data.pdbj.org/pub/pdb/validation_reports/c3/2c3b ftp://data.pdbj.org/pub/pdb/validation_reports/c3/2c3b
ftp://data.pdbj.org/pub/pdb/validation_reports/c3/2c3b Links
Links Assembly
Assembly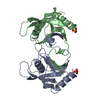
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 0.9999
/ Beamline: X06SA / Wavelength: 0.9999  Processing
Processing MAD / Resolution: 1.85→55.9 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.952 / SU B: 5.037 / SU ML: 0.077 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.115 / ESU R Free: 0.11 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
MAD / Resolution: 1.85→55.9 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.952 / SU B: 5.037 / SU ML: 0.077 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.115 / ESU R Free: 0.11 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. Movie
Movie Controller
Controller












 PDBj
PDBj



