[English] 日本語
 Yorodumi
Yorodumi- PDB-2bao: Solution NMR structure of the myristoylated N-terminal fragment o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2bao | ||||||
|---|---|---|---|---|---|---|---|
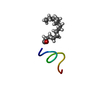
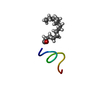
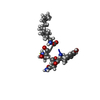
| Title | Solution NMR structure of the myristoylated N-terminal fragment of Arf6 | ||||||
 Components Components | ADP-ribosylation factor 6 | ||||||
 Keywords Keywords | SIGNALING PROTEIN / Myristoyl / N-terminal / Arf6 | ||||||
| Function / homology |  Function and homology information Function and homology informationerythrocyte apoptotic process / maintenance of postsynaptic density structure / protein localization to cleavage furrow / positive regulation of mitotic cytokinetic process / negative regulation of dendrite development / establishment of epithelial cell polarity / regulation of dendritic spine development / regulation of Rac protein signal transduction / negative regulation of protein localization to cell surface / protein localization to endosome ...erythrocyte apoptotic process / maintenance of postsynaptic density structure / protein localization to cleavage furrow / positive regulation of mitotic cytokinetic process / negative regulation of dendrite development / establishment of epithelial cell polarity / regulation of dendritic spine development / regulation of Rac protein signal transduction / negative regulation of protein localization to cell surface / protein localization to endosome / ruffle assembly / positive regulation of keratinocyte migration / regulation of filopodium assembly / positive regulation of focal adhesion disassembly / MET receptor recycling / endocytic recycling / thioesterase binding / Flemming body / filopodium membrane / TBC/RABGAPs / protein localization to cell surface / cortical actin cytoskeleton organization / hepatocyte apoptotic process / positive regulation of actin filament polymerization / cleavage furrow / synaptic vesicle endocytosis / endocytic vesicle / regulation of presynapse assembly / vesicle-mediated transport / ruffle / signaling adaptor activity / small monomeric GTPase / positive regulation of protein secretion / protein localization to plasma membrane / positive regulation of protein localization to plasma membrane / intracellular protein transport / liver development / negative regulation of receptor-mediated endocytosis / positive regulation of neuron projection development / cellular response to nerve growth factor stimulus / recycling endosome membrane / GDP binding / nervous system development / presynapse / Clathrin-mediated endocytosis / G protein activity / early endosome membrane / midbody / cell cortex / cell differentiation / cell adhesion / endosome / postsynapse / cell division / focal adhesion / GTPase activity / GTP binding / glutamatergic synapse / Golgi apparatus / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Gizachew, D. | ||||||
 Citation Citation |  Journal: Febs Lett. / Year: 2006 Journal: Febs Lett. / Year: 2006Title: NMR structural studies of the myristoylated N-terminus of ADP ribosylation factor 6 (Arf6). Authors: Gizachew, D. / Oswald, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2bao.cif.gz 2bao.cif.gz | 46.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2bao.ent.gz pdb2bao.ent.gz | 31.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2bao.json.gz 2bao.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ba/2bao https://data.pdbj.org/pub/pdb/validation_reports/ba/2bao ftp://data.pdbj.org/pub/pdb/validation_reports/ba/2bao ftp://data.pdbj.org/pub/pdb/validation_reports/ba/2bao | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1064.279 Da / Num. of mol.: 1 / Fragment: N-terminal peptide / Source method: obtained synthetically Details: This sequence is the N-terminal peptide of a protein which occurs naturally in Homo sapiens (human). References: UniProt: P62330 |
|---|---|
| #2: Chemical | ChemComp-MYR / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques |
- Sample preparation
Sample preparation
| Details | Contents: 1mM Myristoylated N-terminal Arf6; 5mM acetate buffer Solvent system: 90% H2O/10% D2O |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 500 MHz |
- Processing
Processing
| NMR software | Name:  X-PLOR / Version: NIH-2.9.9 / Developer: Lakowski / Classification: refinement X-PLOR / Version: NIH-2.9.9 / Developer: Lakowski / Classification: refinement |
|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 |
| NMR representative | Selection criteria: lowest energy |
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 12 |
 Movie
Movie Controller
Controller




 PDBj
PDBj










