[English] 日本語
 Yorodumi
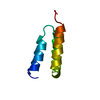
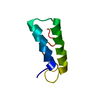
Yorodumi- PDB-1zy6: Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hair... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1zy6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | ||||||
 Components Components | Protegrin 1 | ||||||
 Keywords Keywords | ANTIBIOTIC / beta-hairpin / solid state NMR | ||||||
| Function / homology |  Function and homology information Function and homology informationlipopolysaccharide binding / antimicrobial humoral immune response mediated by antimicrobial peptide / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium / innate immune response / extracellular space Similarity search - Function | ||||||
| Method | SOLID-STATE NMR / Distance geometry; Simulated annealing | ||||||
 Authors Authors | Wu, X. / Mani, R. / Tang, M. / Buffy, J.J. / Waring, A.J. / Sherman, M.A. / Hong, M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2006 Journal: Biochemistry / Year: 2006Title: Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR. Authors: Mani, R. / Tang, M. / Wu, X. / Buffy, J.J. / Waring, A.J. / Sherman, M.A. / Hong, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1zy6.cif.gz 1zy6.cif.gz | 19.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1zy6.ent.gz pdb1zy6.ent.gz | 13.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1zy6.json.gz 1zy6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1zy6_validation.pdf.gz 1zy6_validation.pdf.gz | 242.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1zy6_full_validation.pdf.gz 1zy6_full_validation.pdf.gz | 241.8 KB | Display | |
| Data in XML |  1zy6_validation.xml.gz 1zy6_validation.xml.gz | 1.9 KB | Display | |
| Data in CIF |  1zy6_validation.cif.gz 1zy6_validation.cif.gz | 2.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zy/1zy6 https://data.pdbj.org/pub/pdb/validation_reports/zy/1zy6 ftp://data.pdbj.org/pub/pdb/validation_reports/zy/1zy6 ftp://data.pdbj.org/pub/pdb/validation_reports/zy/1zy6 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2164.679 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: BIOLOGICAL SEQUENCE WITH AMIDATED C-TERMINUS / References: UniProt: P32194 Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||
| NMR details | Text: Solid State NMR was performed at 400.49 MHz for 1H, 376.8 MHz for 19F, 100.72 MHz for 13C, and 40.58 MHz for 15N. The 15N{13C} and 13C{1H} REDOR experiments were carried out on a 1H/13C/15N ...Text: Solid State NMR was performed at 400.49 MHz for 1H, 376.8 MHz for 19F, 100.72 MHz for 13C, and 40.58 MHz for 15N. The 15N{13C} and 13C{1H} REDOR experiments were carried out on a 1H/13C/15N triple resonance MAS probe. Spinning speeds were regulated to 3 Hz using a pneumatic control unit. 13C and 15N chemical shifts were referenced externally to the 13C signal of alpha-Gly at 176.4 ppm on the TMS scale and the 15N signal of N-acetyl-valine at 122 ppm, respectively. The 13C{19F} REDOR experiments, where the nucleus in the bracket is the unobserved dephasing spin, was conducted on a 4-mm magic-angle spinning (MAS) probe equipped with a Bruker HFX unit, which allows simultaneous tuning of 1H and 19F on the 1H channel. |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Pressure: ambient / Temperature: 233 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker DSX 400 / Manufacturer: Bruker / Model: DSX 400 / Field strength: 400 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Distance geometry; Simulated annealing / Software ordinal: 1 Details: Two-spin REDOR curves were simulated using a Fortran program. Three-spin REDOR curves for the C -F experiment were simulated using the SIMPSON program. The input distances and angles in the ...Details: Two-spin REDOR curves were simulated using a Fortran program. Three-spin REDOR curves for the C -F experiment were simulated using the SIMPSON program. The input distances and angles in the three-spin simulation were obtained from model building. The simulations assumed delta-function pulses for all pi pulses. Models that were potentially consistent with all the experimentally measured distances were created using MODELLER. In addition to the REDOR-based restraints, the input file included a restraint to preserve the intramolecular hydrogen bond ladder of each monomer, and a restraint to maintain monomer symmetry. | ||||||||||||
| NMR ensemble | Conformer selection criteria: least violations / Conformers calculated total number: 100 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller









 PDBj
PDBj
 MODELLER
MODELLER