[English] 日本語
 Yorodumi
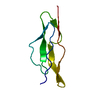
Yorodumi- PDB-1yuf: TYPE ALPHA TRANSFORMING GROWTH FACTOR, NMR, 16 MODELS WITHOUT ENE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1yuf | ||||||
|---|---|---|---|---|---|---|---|
| Title | TYPE ALPHA TRANSFORMING GROWTH FACTOR, NMR, 16 MODELS WITHOUT ENERGY MINIMIZATION | ||||||
 Components Components | TRANSFORMING GROWTH FACTOR ALPHA | ||||||
 Keywords Keywords | GROWTH FACTOR / EGF-LIKE DOMAIN STRUCTURE | ||||||
| Function / homology |  Function and homology information Function and homology informationhepatocyte proliferation / transmembrane receptor protein tyrosine kinase activator activity / Cargo concentration in the ER / COPII-mediated vesicle transport / epidermal growth factor receptor binding / Inhibition of Signaling by Overexpressed EGFR / EGFR interacts with phospholipase C-gamma / ERBB2-EGFR signaling pathway / Signaling by EGFR / positive regulation of cell division ...hepatocyte proliferation / transmembrane receptor protein tyrosine kinase activator activity / Cargo concentration in the ER / COPII-mediated vesicle transport / epidermal growth factor receptor binding / Inhibition of Signaling by Overexpressed EGFR / EGFR interacts with phospholipase C-gamma / ERBB2-EGFR signaling pathway / Signaling by EGFR / positive regulation of cell division / mammary gland alveolus development / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / GAB1 signalosome / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / GRB2 events in EGFR signaling / SHC1 events in EGFR signaling / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / positive regulation of epithelial cell proliferation / growth factor activity / clathrin-coated endocytic vesicle membrane / EGFR downregulation / ER to Golgi transport vesicle membrane / epidermal growth factor receptor signaling pathway / Constitutive Signaling by Aberrant PI3K in Cancer / PIP3 activates AKT signaling / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / RAF/MAP kinase cascade / cytoplasmic vesicle / angiogenesis / basolateral plasma membrane / Estrogen-dependent gene expression / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / positive regulation of MAPK cascade / intracellular signal transduction / receptor ligand activity / positive regulation of cell population proliferation / endoplasmic reticulum membrane / perinuclear region of cytoplasm / cell surface / extracellular space / extracellular region / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Moy, F.J. / Montelione, G.T. / Scheraga, H.A. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1993 Journal: Biochemistry / Year: 1993Title: Solution structure of human type-alpha transforming growth factor determined by heteronuclear NMR spectroscopy and refined by energy minimization with restraints. Authors: Moy, F.J. / Li, Y.C. / Rauenbuehler, P. / Winkler, M.E. / Scheraga, H.A. / Montelione, G.T. #1:  Journal: J.Biomol.NMR / Year: 1995 Journal: J.Biomol.NMR / Year: 1995Title: Crankshaft Motions of the Polypeptide Backbone in Molecular Dynamics Simulations of Human Type-Alpha Transforming Growth Factor Authors: Fadel, A.R. / Jin, D.Q. / Montelione, G.T. / Levy, R.M. #2:  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: Human Type-Alpha Transforming Growth Factor Undergoes Slow Conformational Exchange between Multiple Backbone Conformations as Characterized by Nitrogen-15 Relaxation Measurements Authors: Li, Y.C. / Montelione, G.T. #3:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1989 Journal: Proc.Natl.Acad.Sci.USA / Year: 1989Title: Sequence-Specific 1H-NMR Assignments and Identification of Two Small Antiparallel Beta-Sheets in the Solution Structure of Recombinant Human Transforming Growth Factor Alpha Authors: Montelione, G.T. / Winkler, M.E. / Burton, L.E. / Rinderknecht, E. / Sporn, M.B. / Wagner, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1yuf.cif.gz 1yuf.cif.gz | 232.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1yuf.ent.gz pdb1yuf.ent.gz | 190.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1yuf.json.gz 1yuf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yu/1yuf https://data.pdbj.org/pub/pdb/validation_reports/yu/1yuf ftp://data.pdbj.org/pub/pdb/validation_reports/yu/1yuf ftp://data.pdbj.org/pub/pdb/validation_reports/yu/1yuf | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 5560.246 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: HUMAN TGF-ALPHA 1YUF / Plasmid: PTE5 / Gene (production host): HUMAN TGF-ALPHA 1YUF / Production host: Homo sapiens (human) / Gene: HUMAN TGF-ALPHA 1YUF / Plasmid: PTE5 / Gene (production host): HUMAN TGF-ALPHA 1YUF / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| NMR software | Name: DISMAN / Developer: BRAUN,GO / Classification: refinement |
|---|---|
| NMR ensemble | Conformers submitted total number: 16 |
 Movie
Movie Controller
Controller








 PDBj
PDBj









