[English] 日本語
 Yorodumi
Yorodumi- PDB-1ypa: DIRECT OBSERVATION OF BETTER HYDRATION AT THE N-TERMINUS OF AN AL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ypa | ||||||
|---|---|---|---|---|---|---|---|
| Title | DIRECT OBSERVATION OF BETTER HYDRATION AT THE N-TERMINUS OF AN ALPHA-HELIX WITH GLYCINE RATHER THAN ALANINE AS N-CAP | ||||||
 Components Components | CHYMOTRYPSIN INHIBITOR 2 | ||||||
 Keywords Keywords | PROTEINASE INHIBITOR(CHYMOTRYPSIN) | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Harpaz, Y. / Elmasry, N. / Fersht, A.R. / Henrick, K. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 1994 Journal: Proc.Natl.Acad.Sci.USA / Year: 1994Title: Direct observation of better hydration at the N terminus of an alpha-helix with glycine rather than alanine as the N-cap residue. Authors: Harpaz, Y. / Elmasry, N. / Fersht, A.R. / Henrick, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ypa.cif.gz 1ypa.cif.gz | 23.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ypa.ent.gz pdb1ypa.ent.gz | 14.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ypa.json.gz 1ypa.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ypa_validation.pdf.gz 1ypa_validation.pdf.gz | 356.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ypa_full_validation.pdf.gz 1ypa_full_validation.pdf.gz | 356.9 KB | Display | |
| Data in XML |  1ypa_validation.xml.gz 1ypa_validation.xml.gz | 2.4 KB | Display | |
| Data in CIF |  1ypa_validation.cif.gz 1ypa_validation.cif.gz | 3.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yp/1ypa https://data.pdbj.org/pub/pdb/validation_reports/yp/1ypa ftp://data.pdbj.org/pub/pdb/validation_reports/yp/1ypa ftp://data.pdbj.org/pub/pdb/validation_reports/yp/1ypa | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 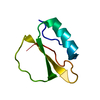
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 7181.550 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.92 Å3/Da / Density % sol: 35.99 % |
|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop |
-Data collection
| Reflection | *PLUS Highest resolution: 2 Å / Lowest resolution: 14.5 Å / Num. obs: 3689 / % possible obs: 93 % / Num. measured all: 14561 / Rmerge(I) obs: 0.077 |
|---|---|
| Reflection shell | *PLUS % possible obs: 96.4 % / Rmerge(I) obs: 0.129 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.178 / Highest resolution: 2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Rfactor obs: 0.178 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 11.6 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: p_angle_d / Dev ideal: 0.033 |
 Movie
Movie Controller
Controller










 PDBj
PDBj
