| 登録情報 | データベース: PDB / ID: 1y8o
|
|---|
| タイトル | Crystal structure of the PDK3-L2 complex |
|---|
 要素 要素 | - Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex
- [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3
|
|---|
 キーワード キーワード | TRANSFERASE / pyruvate dehydrogenase kinase 3 / lipoyl-bearing domain / protein-protein complex |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
hypoxia-inducible factor-1alpha signaling pathway / PDH complex synthesizes acetyl-CoA from PYR / [pyruvate dehydrogenase (acetyl-transferring)] kinase / regulation of acetyl-CoA biosynthetic process from pyruvate / pyruvate dehydrogenase (acetyl-transferring) kinase activity / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / acetyl-CoA biosynthetic process from pyruvate / Regulation of pyruvate dehydrogenase (PDH) complex / Protein lipoylation ...hypoxia-inducible factor-1alpha signaling pathway / PDH complex synthesizes acetyl-CoA from PYR / [pyruvate dehydrogenase (acetyl-transferring)] kinase / regulation of acetyl-CoA biosynthetic process from pyruvate / pyruvate dehydrogenase (acetyl-transferring) kinase activity / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / acetyl-CoA biosynthetic process from pyruvate / Regulation of pyruvate dehydrogenase (PDH) complex / Protein lipoylation / pyruvate dehydrogenase complex / regulation of reactive oxygen species metabolic process / cellular response to fatty acid / Signaling by Retinoic Acid / regulation of glucose metabolic process / tricarboxylic acid cycle / peroxisome proliferator activated receptor signaling pathway / cellular response to glucose stimulus / glucose metabolic process / peptidyl-serine phosphorylation / protein kinase activity / mitochondrial matrix / intracellular membrane-bounded organelle / protein serine/threonine kinase activity / nucleolus / mitochondrion / ATP binding / identical protein binding類似検索 - 分子機能 Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex / Dihydrolipoamide acetyltransferase/Pyruvate dehydrogenase protein X component / Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain / Branched-chain alpha-ketoacid dehydrogenase kinase/Pyruvate dehydrogenase kinase, N-terminal / Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain superfamily / PDK/BCKDK protein kinase / Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase / Peripheral subunit-binding domain / e3 binding domain / Peripheral subunit-binding (PSBD) domain profile. ...Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex / Dihydrolipoamide acetyltransferase/Pyruvate dehydrogenase protein X component / Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain / Branched-chain alpha-ketoacid dehydrogenase kinase/Pyruvate dehydrogenase kinase, N-terminal / Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain superfamily / PDK/BCKDK protein kinase / Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase / Peripheral subunit-binding domain / e3 binding domain / Peripheral subunit-binding (PSBD) domain profile. / E3-binding domain superfamily / 2-oxo acid dehydrogenase, lipoyl-binding site / 2-oxo acid dehydrogenases acyltransferase component lipoyl binding site. / 2-oxoacid dehydrogenase acyltransferase, catalytic domain / 2-oxoacid dehydrogenases acyltransferase (catalytic domain) / RNA polymerase II/Efflux pump adaptor protein, barrel-sandwich hybrid domain / Butyryl-CoA Dehydrogenase, subunit A; domain 3 / Biotin-requiring enzyme / Biotinyl/lipoyl domain profile. / Biotin/lipoyl attachment / Single hybrid motif / Chloramphenicol acetyltransferase-like domain superfamily / Histidine kinase domain / Histidine kinase domain profile. / Histidine kinase-like ATPase, C-terminal domain / Heat Shock Protein 90 / Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase / Histidine kinase-like ATPases / Histidine kinase/HSP90-like ATPase / Histidine kinase/HSP90-like ATPase superfamily / OB fold (Dihydrolipoamide Acetyltransferase, E2P) / Up-down Bundle / Beta Barrel / 2-Layer Sandwich / Mainly Beta / Mainly Alpha / Alpha Beta類似検索 - ドメイン・相同性 ADENOSINE-5'-DIPHOSPHATE / : / DIHYDROLIPOIC ACID / Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial / [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 3, mitochondrial類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.48 Å 分子置換 / 解像度: 2.48 Å |
|---|
 データ登録者 データ登録者 | Kato, M. / Chuang, J.L. / Wynn, R.M. / Chuang, D.T. |
|---|
 引用 引用 |  ジャーナル: Embo J. / 年: 2005 ジャーナル: Embo J. / 年: 2005
タイトル: Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
著者: Kato, M. / Chuang, J.L. / Tso, S.C. / Wynn, R.M. / Chuang, D.T. |
|---|
| 履歴 | | 登録 | 2004年12月13日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2005年5月24日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2008年4月30日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2011年7月13日 | Group: Derived calculations / Version format compliance |
|---|
| 改定 1.3 | 2011年10月5日 | Group: Non-polymer description |
|---|
| 改定 1.4 | 2023年8月23日 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_atom_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.48 Å
分子置換 / 解像度: 2.48 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Embo J. / 年: 2005
ジャーナル: Embo J. / 年: 2005 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1y8o.cif.gz
1y8o.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1y8o.ent.gz
pdb1y8o.ent.gz PDB形式
PDB形式 1y8o.json.gz
1y8o.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 1y8o_validation.pdf.gz
1y8o_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 1y8o_full_validation.pdf.gz
1y8o_full_validation.pdf.gz 1y8o_validation.xml.gz
1y8o_validation.xml.gz 1y8o_validation.cif.gz
1y8o_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/y8/1y8o
https://data.pdbj.org/pub/pdb/validation_reports/y8/1y8o ftp://data.pdbj.org/pub/pdb/validation_reports/y8/1y8o
ftp://data.pdbj.org/pub/pdb/validation_reports/y8/1y8o リンク
リンク 集合体
集合体

 要素
要素 Homo sapiens (ヒト) / 遺伝子: PDK3 / プラスミド: pTrcHisB / 生物種 (発現宿主): Escherichia coli / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: PDK3 / プラスミド: pTrcHisB / 生物種 (発現宿主): Escherichia coli / 発現宿主: 
 Homo sapiens (ヒト) / 遺伝子: DLAT, DLTA / プラスミド: pTrcHisB / 生物種 (発現宿主): Escherichia coli / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: DLAT, DLTA / プラスミド: pTrcHisB / 生物種 (発現宿主): Escherichia coli / 発現宿主: 



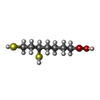





 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 19-ID / 波長: 0.97 Å
/ ビームライン: 19-ID / 波長: 0.97 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj








