[English] 日本語
 Yorodumi
Yorodumi- PDB-1y7k: NMR structure family of Human Agouti Signalling Protein (80-132: ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1y7k | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | ||||||
 Components Components | Agouti Signaling Protein | ||||||
 Keywords Keywords | SIGNALING PROTEIN / Melanocortin / inhibitor / cystine knot / GPCR / endogenous antagonist / inverse agonist | ||||||
| Function / homology |  Function and homology information Function and homology informationmelanocortin receptor binding / negative regulation of melanin biosynthetic process / type 3 melanocortin receptor binding / type 4 melanocortin receptor binding / type 1 melanocortin receptor binding / receptor antagonist activity / positive regulation of melanin biosynthetic process / adult feeding behavior / melanin biosynthetic process / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway ...melanocortin receptor binding / negative regulation of melanin biosynthetic process / type 3 melanocortin receptor binding / type 4 melanocortin receptor binding / type 1 melanocortin receptor binding / receptor antagonist activity / positive regulation of melanin biosynthetic process / adult feeding behavior / melanin biosynthetic process / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / melanosome organization / melanosome transport / neuropeptide hormone activity / epigenetic programming in the zygotic pronuclei / hormone-mediated signaling pathway / generation of precursor metabolites and energy / cell-cell signaling / signaling receptor binding / signal transduction / extracellular space Similarity search - Function | ||||||
| Method | SOLUTION NMR / simulated annealing, torsion angle dynamics | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | McNulty, J.C. / Jackson, P.J. / Thompson, D.A. / Chai, B. / Gantz, I. / Barsh, G.S. / Dawson, P.E. / Millhauser, G.L. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2005 Journal: J.Mol.Biol. / Year: 2005Title: Structures of the agouti signaling protein. Authors: McNulty, J.C. / Jackson, P.J. / Thompson, D.A. / Chai, B. / Gantz, I. / Barsh, G.S. / Dawson, P.E. / Millhauser, G.L. #1:  Journal: Biochemistry / Year: 2001 Journal: Biochemistry / Year: 2001Title: High-resolution NMR structure of the chemically-synthesized melanocortin receptor binding domain AGRP(87-132) of the agouti-related protein Authors: McNulty, J.C. / Thompson, D.A. / Bolin, K.A. / Wilken, J. / Barsh, G.S. / Millhauser, G.L. #2:  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity Authors: Jackson, P.J. / McNulty, J.C. / Yang, Y.K. / Thompson, D.A. / Chai, B. / Gantz, I. / Barsh, G.S. / Millhauser, G.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1y7k.cif.gz 1y7k.cif.gz | 240.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1y7k.ent.gz pdb1y7k.ent.gz | 197.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1y7k.json.gz 1y7k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y7/1y7k https://data.pdbj.org/pub/pdb/validation_reports/y7/1y7k ftp://data.pdbj.org/pub/pdb/validation_reports/y7/1y7k ftp://data.pdbj.org/pub/pdb/validation_reports/y7/1y7k | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 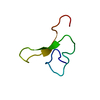
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 5823.997 Da / Num. of mol.: 1 / Fragment: residies 80-132 / Mutation: Q115Y, S124Y / Source method: obtained synthetically Details: Sequence occurs naturally in humans, genes ASIP, AGTI, AGTIL, ASP References: UniProt: P42127 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques, in conjunction with a semi-automated assignment protocol. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, torsion angle dynamics / Software ordinal: 1 Details: This structure family represents one of two major conformers present in solution. The two conformers arise from the cis-trans proline isomerization of the Ala104-Pro105 peptide bond. | ||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 1000 / Conformers submitted total number: 21 Distance constraint violation method: From the original family of twenty, the representative conformer had the lowest target function and was subjected to further minimization in water. The remaining ...Distance constraint violation method: From the original family of twenty, the representative conformer had the lowest target function and was subjected to further minimization in water. The remaining structures were obtained without explicit inclusion of solvent. |
 Movie
Movie Controller
Controller









 PDBj
PDBj CYANA
CYANA