[English] 日本語
 Yorodumi
Yorodumi- PDB-1wqy: X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1wqy | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
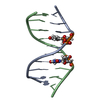
| Title | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / B-DNA / HYDRATION / HYDROGEN | Function / homology | DEUTERATED WATER / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å  Authors AuthorsArai, S. / Chatake, T. / Ohhara, T. / Kurihara, K. / Tanaka, I. / Suzuki, N. / Fujimoto, Z. / Mizuno, H. / Niimura, N. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2005 Journal: Nucleic Acids Res. / Year: 2005Title: Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements Authors: Arai, S. / Chatake, T. / Ohhara, T. / Kurihara, K. / Tanaka, I. / Suzuki, N. / Fujimoto, Z. / Mizuno, H. / Niimura, N. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1wqy.cif.gz 1wqy.cif.gz | 21.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1wqy.ent.gz pdb1wqy.ent.gz | 13.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1wqy.json.gz 1wqy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wq/1wqy https://data.pdbj.org/pub/pdb/validation_reports/wq/1wqy ftp://data.pdbj.org/pub/pdb/validation_reports/wq/1wqy ftp://data.pdbj.org/pub/pdb/validation_reports/wq/1wqy | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1wqzC  167dS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3044.017 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | ChemComp-DOD / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.12 % | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 279 K / Method: batch (d2o) / pH: 7 Details: 30% MPD, 0.1M MAGNESIUM CHLORIDE, 0.1M SODIUM CACODYLATE, 0.002M DNA, pD 6.6, pH 7.0, BATCH (D2O), temperature 279K | ||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 93 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 0.71 / Wavelength: 0.71 Å / Beamline: BL41XU / Wavelength: 0.71 / Wavelength: 0.71 Å |
| Detector | Detector: IMAGE PLATE / Date: Jun 25, 2003 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.71 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→50 Å / Num. all: 8528 / Num. obs: 8451 / % possible obs: 99.1 % / Observed criterion σ(I): 0 / Redundancy: 9.27 % / Biso Wilson estimate: 22.3 Å2 / Rmerge(I) obs: 0.025 |
| Reflection shell | Resolution: 1.6→1.66 Å / Redundancy: 5.25 % / Rmerge(I) obs: 0.108 / Num. unique all: 828 / % possible all: 97.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 167D Resolution: 2→50 Å / Cross valid method: THROUGHOUT / σ(F): 0 Details: LIMITING RESOLUTION ESTIMATED FROM DATA COLLECTION STATISTICS WAS 1.6 ANGSTROMS. HOWEVER, THE STRUCTURAL REFINEMENT PROCESSING WAS COMPUTED BY USING REFLECTIONS 2.0 - 50.0 ANGSTROMS BECAUSE ...Details: LIMITING RESOLUTION ESTIMATED FROM DATA COLLECTION STATISTICS WAS 1.6 ANGSTROMS. HOWEVER, THE STRUCTURAL REFINEMENT PROCESSING WAS COMPUTED BY USING REFLECTIONS 2.0 - 50.0 ANGSTROMS BECAUSE THE BIN R VALUE AND BIN FREE R VALUE OF RESOLUTION RANGE HIGHER THAN 2.0 ANGSTROMS WERE VERY HIGH.
| |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→50 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.09 Å / Rfactor Rfree error: 0.048
|
 Movie
Movie Controller
Controller








 PDBj
PDBj





