[English] 日本語
 Yorodumi
Yorodumi- PDB-1w31: YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1w31 | ||||||
|---|---|---|---|---|---|---|---|
| Title | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | ||||||
 Components Components | DELTA-AMINOLEVULINIC ACID DEHYDRATASE | ||||||
 Keywords Keywords | LYASE / DEHYDRATASE / ALDOLASE / TIM BARREL / TETRAPYRROLE SYNTHESIS / HEME BIOSYNTHESIS / ZINC | ||||||
| Function / homology |  Function and homology information Function and homology informationHeme biosynthesis / porphobilinogen synthase / porphobilinogen synthase activity / protoporphyrinogen IX biosynthetic process / heme biosynthetic process / Neutrophil degranulation / zinc ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Erskine, P.T. / Coates, L. / Newbold, R. / Brindley, A.A. / Stauffer, F. / Beaven, G.D.E. / Gill, R. / Wood, S.P. / Warren, M.J. / Cooper, J.B. ...Erskine, P.T. / Coates, L. / Newbold, R. / Brindley, A.A. / Stauffer, F. / Beaven, G.D.E. / Gill, R. / Wood, S.P. / Warren, M.J. / Cooper, J.B. / Shoolingin-Jordan, P.M. / Neier, R. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2005 Journal: Acta Crystallogr.,Sect.D / Year: 2005Title: Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid Authors: Erskine, P.T. / Coates, L. / Newbold, R. / Brindley, A.A. / Stauffer, F. / Beaven, G.D.E. / Gill, R. / Coker, A. / Wood, S.P. / Warren, M.J. / Shoolingin-Jordan, P.M. / Neier, R. / Cooper, J.B. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 11-STRANDED BARREL THIS IS REPRESENTED BY A 12-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1w31.cif.gz 1w31.cif.gz | 86.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1w31.ent.gz pdb1w31.ent.gz | 65.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1w31.json.gz 1w31.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w3/1w31 https://data.pdbj.org/pub/pdb/validation_reports/w3/1w31 ftp://data.pdbj.org/pub/pdb/validation_reports/w3/1w31 ftp://data.pdbj.org/pub/pdb/validation_reports/w3/1w31 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ylvS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | x 8
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 37785.160 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: COMPLEX WITH 5-HYDROXYLAEVULINIC ACID Source: (gene. exp.)  Strain: NS1(JM109/PNS1) / Production host:  |
|---|---|
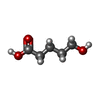
| #2: Chemical | ChemComp-SHO / |
| #3: Chemical | ChemComp-ZN / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.91 Å3/Da / Density % sol: 57 % |
|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 8 Details: ENZYME CONCENTRATION 1 MG/ML, PH 7.0 - 8.5, BUFFER 0.2 M TRIS-HCL, PRECIPITANT PEG 6000 (<10%), 70 MICROMOLAR ZINC SULPHATE, 6 MM BETA-MERCAPTOETHANOL, HANGING DROPS AS FOR PDB ENTRY 1AW5 WITH 10 MM 5-HYDROXYLAEVULINIC ACID IN DROP. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID29 / Wavelength: 0.911662 / Beamline: ID29 / Wavelength: 0.911662 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Feb 16, 2002 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.911662 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→49 Å / Num. obs: 1067852 / % possible obs: 100 % / Redundancy: 11.9 % / Rmerge(I) obs: 0.058 / Net I/σ(I): 8 |
| Reflection shell | Resolution: 1.9→2 Å / Redundancy: 10 % / Rmerge(I) obs: 0.419 / Mean I/σ(I) obs: 1.7 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1YLV Resolution: 1.9→44.28 Å / Cross valid method: FREE R-VALUE / σ(F): 0
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→44.28 Å
| |||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.1895 / Rfactor Rfree: 0.2493 / Rfactor Rwork: 0.1895 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller






















 PDBj
PDBj