+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1vtg | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA/ANTIBIOTIC / BISINTERCALATOR / DEPSIPEPTIDE / QUINOXALINE / ANTIBIOTIC / ANTITUMOR / DNA-ANTIBIOTIC COMPLEX | ||||||
| Function / homology | TRIOSTIN A / 2-CARBOXYQUINOXALINE / : / DNA Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) Streptomyces sp. (bacteria) Streptomyces sp. (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.67 Å X-RAY DIFFRACTION / Resolution: 1.67 Å | ||||||
 Authors Authors | Wang, A.H.-J. / Ughetto, G. / Quigley, G.J. / Hakoshima, T. / Van Der Marel, G.A. / Van Boom, J.H. / Rich, A. | ||||||
 Citation Citation |  Journal: Science / Year: 1984 Journal: Science / Year: 1984Title: The molecular structure of a DNA-triostin A complex. Authors: Wang, A.H. / Ughetto, G. / Quigley, G.J. / Hakoshima, T. / van der Marel, G.A. / van Boom, J.H. / Rich, A. #1: Journal: Nucleic Acids Res. / Year: 1985 Title: A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Authors: Ughetto, G. / Wang, A.H. / Quigley, G.J. / van der Marel, G.A. / van Boom, J.H. / Rich, A. #2:  Journal: J. Biomol. Struct. Dyn. / Year: 1986 Journal: J. Biomol. Struct. Dyn. / Year: 1986Title: Interactions of quinoxaline antibiotic and DNA: the molecular structure of a triostin A-d(GCGTACGC) complex. Authors: Wang, A.H. / Ughetto, G. / Quigley, G.J. / Rich, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1vtg.cif.gz 1vtg.cif.gz | 17.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1vtg.ent.gz pdb1vtg.ent.gz | 10.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1vtg.json.gz 1vtg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1vtg_validation.pdf.gz 1vtg_validation.pdf.gz | 367.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1vtg_full_validation.pdf.gz 1vtg_full_validation.pdf.gz | 381.8 KB | Display | |
| Data in XML |  1vtg_validation.xml.gz 1vtg_validation.xml.gz | 4.6 KB | Display | |
| Data in CIF |  1vtg_validation.cif.gz 1vtg_validation.cif.gz | 5.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vt/1vtg https://data.pdbj.org/pub/pdb/validation_reports/vt/1vtg ftp://data.pdbj.org/pub/pdb/validation_reports/vt/1vtg ftp://data.pdbj.org/pub/pdb/validation_reports/vt/1vtg | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 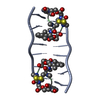
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 1809.217 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) | ||
|---|---|---|---|
| #2: Protein/peptide | 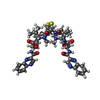  Type: Cyclic depsipeptide / Class: Anticancer / Mass: 794.982 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Cyclic depsipeptide / Class: Anticancer / Mass: 794.982 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE ...Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. Source: (synth.)  Streptomyces sp. (bacteria) / References: NOR: NOR01129, TRIOSTIN A Streptomyces sp. (bacteria) / References: NOR: NOR01129, TRIOSTIN A | ||
| #3: Chemical | 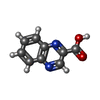  Type: Cyclic depsipeptide / Class: Anticancer / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2 Type: Cyclic depsipeptide / Class: Anticancer / Mass: 174.156 Da / Num. of mol.: 2 / Source method: obtained synthetically / Formula: C9H6N2O2Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE ...Details: TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE. BICYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI, AND A DISULPHIDE BOND BETWEEN RESIDUES 3 AND 7. THE TWO QUINOXALINE CHROMOPHORES ARE LINKED TO THE D-SERINE RESIDUES, RESIDUES 1 AND 5. References: TRIOSTIN A Compound details | TRIOSTIN IS A BICYCLIC OCTADEPSIPEPTIDE, A MEMBER OF THE QUINOXALINE CLASS OF ANTIBIOTICS. HERE, ...TRIOSTIN IS A BICYCLIC OCTADEPSIP | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.14 Å3/Da / Density % sol: 70.28 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion / pH: 7 / Details: pH 7.00, VAPOR DIFFUSION | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 257 K |
|---|---|
| Detector | Type: NICOLET P3 / Detector: DIFFRACTOMETER |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Num. obs: 2995 / Observed criterion σ(I): 1 |
- Processing
Processing
| Software | Name: NUCLSQ / Classification: refinement | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.189 / Highest resolution: 1.67 Å Details: COORDINATES ARE REPORTED IN THE JRNL REFERENCE PAPER AS A COMPARISON. | ||||||||||||
| Refine Biso | Class: ALL ATOMS / Details: TR / Treatment: isotropic | ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.67 Å
|
 Movie
Movie Controller
Controller









 PDBj
PDBj