登録情報 データベース : PDB / ID : 1vqrタイトル Crystal structure of a virulence factor (cj0248) from campylobacter jejuni subsp. jejuni at 2.25 A resolution hypothetical protein Cj0248 キーワード / / / / / / 機能・相同性 ドメイン・相同性 構成要素
/ / / / / / / 生物種 Campylobacter jejuni subsp. jejuni NCTC 11168 (カンピロバクター)手法 / / / 解像度 : 2.25 Å データ登録者 Joint Center for Structural Genomics (JCSG) ジャーナル : Proteins / 年 : 2006タイトル : Crystal structure of virulence factor CJ0248 from Campylobacter jejuni at 2.25 A resolution reveals a new fold.著者: Xu, Q. / Schwarzenbacher, R. / McMullan, D. / Abdubek, P. / Agarwalla, S. / Ambing, E. / Axelrod, H. / Biorac, T. / Canaves, J.M. / Chiu, H.J. / Deacon, A.M. / DiDonato, M. / Elsliger, M.A. / ... 著者 : Xu, Q. / Schwarzenbacher, R. / McMullan, D. / Abdubek, P. / Agarwalla, S. / Ambing, E. / Axelrod, H. / Biorac, T. / Canaves, J.M. / Chiu, H.J. / Deacon, A.M. / DiDonato, M. / Elsliger, M.A. / Godzik, A. / Grittini, C. / Grzechnik, S.K. / Hale, J. / Hampton, E. / Han, G.W. / Haugen, J. / Hornsby, M. / Jaroszewski, L. / Klock, H.E. / Koesema, E. / Kreusch, A. / Kuhn, P. / Lesley, S.A. / Miller, M.D. / Moy, K. / Nigoghossian, E. / Paulsen, J. / Quijano, K. / Reyes, R. / Rife, C. / Spraggon, G. / Stevens, R.C. / van den Bedem, H. / Velasquez, J. / White, A. / Wolf, G. / Hodgson, K.O. / Wooley, J. / Wilson, I.A. 履歴 登録 2004年12月17日 登録サイト / 処理サイト 改定 1.0 2004年12月28日 Provider / タイプ 改定 1.1 2008年4月27日 Group 改定 1.2 2011年7月13日 Group / Version format compliance改定 1.3 2017年10月4日 Group / カテゴリ 改定 1.4 2017年10月25日 Group / カテゴリ 改定 1.5 2023年1月25日 Group / Derived calculations / カテゴリ / struct_conn / struct_ref_seq_difItem _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag / _struct_ref_seq_dif.details 改定 1.6 2024年10月9日 Group / Refinement description / Structure summaryカテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / pdbx_entry_details / pdbx_modification_feature / struct_ncs_dom_lim Item / _struct_ncs_dom_lim.end_auth_comp_id
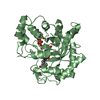
すべて表示 表示を減らす Remark 300 BIOMOLECULE: 1, 2, 3, 4 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF ... BIOMOLECULE: 1, 2, 3, 4 THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 4 CHAIN(S). SEE REMARK 350 FOR INFORMATION ON GENERATING THE BIOLOGICAL MOLECULE(S). SIZE EXCLUSION CHROMATOGRAPHY (SEC) AND MULTI-ANGLE LIGHT SCATTERING (MALS) SUPPORT THE ASSIGNMENT OF THE BIOLOGICAL OLIGOMERIZATION STATE AS A MONOMER.
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Campylobacter jejuni subsp. jejuni NCTC 11168 (カンピロバクター)
Campylobacter jejuni subsp. jejuni NCTC 11168 (カンピロバクター) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  単波長異常分散 / 解像度: 2.25 Å
単波長異常分散 / 解像度: 2.25 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Proteins / 年: 2006
ジャーナル: Proteins / 年: 2006 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1vqr.cif.gz
1vqr.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1vqr.ent.gz
pdb1vqr.ent.gz PDB形式
PDB形式 1vqr.json.gz
1vqr.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/vq/1vqr
https://data.pdbj.org/pub/pdb/validation_reports/vq/1vqr ftp://data.pdbj.org/pub/pdb/validation_reports/vq/1vqr
ftp://data.pdbj.org/pub/pdb/validation_reports/vq/1vqr リンク
リンク 集合体
集合体




 ムービー
ムービー コントローラー
コントローラー












 PDBj
PDBj