| 登録情報 | データベース: PDB / ID: 1vje
|
|---|
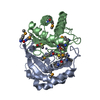
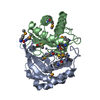
| タイトル | Crystal structure of a autoinducer-2 synthesis protein with bound selenomethionine |
|---|
 要素 要素 | Autoinducer-2 production protein LuxS |
|---|
 キーワード キーワード | HYDROLASE / structural genomics |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
S-ribosylhomocysteine lyase / S-ribosylhomocysteine lyase activity / L-methionine salvage from S-adenosylmethionine / quorum sensing / iron ion binding / cytosol類似検索 - 分子機能 S-ribosylhomocysteinase (LuxS) / S-ribosylhomocysteinase (LuxS) / S-ribosylhomocysteinase (LuxS) superfamily / S-Ribosylhomocysteinase (LuxS) / Metalloenzyme, LuxS/M16 peptidase-like / Gyrase A; domain 2 / 2-Layer Sandwich / Alpha Beta類似検索 - ドメイン・相同性 SELENOMETHIONINE / S-ribosylhomocysteine lyase類似検索 - 構成要素 |
|---|
| 生物種 |  Deinococcus radiodurans (放射線耐性) Deinococcus radiodurans (放射線耐性) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.64 Å 分子置換 / 解像度: 1.64 Å |
|---|
 データ登録者 データ登録者 | Structural GenomiX |
|---|
 引用 引用 |  ジャーナル: Structure / 年: 2001タイトル ジャーナル: Structure / 年: 2001タイトル: A Structural Genomics Approach to the Study of Quorum Sensing: Crystal Structures of Three LuxS Orthologs 著者: Lewis, H.A. / Furlong, E.B. / Laubert, B. / Eroshkina, G.A. / Batiyenko, Y. / Adams, J.M. / Bergseid, M.G. / Marsh, C.D. / Peat, T.S. / Sanderson, W.E. / Sauder, J.M. / Buchanan, S.G.#1:  ジャーナル: Proteins / 年: 2005タイトル ジャーナル: Proteins / 年: 2005タイトル: Structural analysis of a set of proteins resulting from a bacterial genomics project 著者: Badger, J. / Sauder, J.M. / Adams, J.M. / Antonysamy, S. / Bain, K. / Bergseid, M.G. / Buchanan, S.G. / Buchanan, M.D. / Batiyenko, Y. / Christopher, J.A. / Emtage, S. / Eroshkina, A. / Feil, ...著者: Badger, J. / Sauder, J.M. / Adams, J.M. / Antonysamy, S. / Bain, K. / Bergseid, M.G. / Buchanan, S.G. / Buchanan, M.D. / Batiyenko, Y. / Christopher, J.A. / Emtage, S. / Eroshkina, A. / Feil, I. / Furlong, E.B. / Gajiwala, K.S. / Gao, X. / He, D. / Hendle, J. / Huber, A. / Hoda, K. / Kearins, P. / Kissinger, C. / Laubert, B. / Lewis, H.A. / Lin, J. / Loomis, K. / Lorimer, D. / Louie, G. / Maletic, M. / Marsh, C.D. / Miller, I. / Molinari, J. / Muller-Dieckmann, H.J. / Newman, J.M. / Noland, B.W. / Pagarigan, B. / Park, F. / Peat, T.S. / Post, K.W. / Radojicic, S. / Ramos, A. / Romero, R. / Rutter, M.E. / Sanderson, W.E. / Schwinn, K.D. / Tresser, J. / Winhoven, J. / Wright, T.A. / Wu, L. / Xu, J. / Harris, T.J. |
|---|
| 履歴 | | 登録 | 2004年2月3日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2004年2月10日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2008年4月26日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2011年7月13日 | Group: Version format compliance |
|---|
| 改定 1.3 | 2017年10月4日 | Group: Refinement description / カテゴリ: software / Item: _software.name |
|---|
| 改定 1.4 | 2023年12月27日 | Group: Data collection / Database references / Derived calculations
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn / struct_ref_seq_dif / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
| 改定 1.5 | 2024年11月6日 | Group: Structure summary
カテゴリ: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Deinococcus radiodurans (放射線耐性)
Deinococcus radiodurans (放射線耐性) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.64 Å
分子置換 / 解像度: 1.64 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Structure / 年: 2001
ジャーナル: Structure / 年: 2001 ジャーナル: Proteins / 年: 2005
ジャーナル: Proteins / 年: 2005 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1vje.cif.gz
1vje.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1vje.ent.gz
pdb1vje.ent.gz PDB形式
PDB形式 1vje.json.gz
1vje.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/vj/1vje
https://data.pdbj.org/pub/pdb/validation_reports/vj/1vje ftp://data.pdbj.org/pub/pdb/validation_reports/vj/1vje
ftp://data.pdbj.org/pub/pdb/validation_reports/vj/1vje リンク
リンク 集合体
集合体
 要素
要素 Deinococcus radiodurans (放射線耐性)
Deinococcus radiodurans (放射線耐性)
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 32-ID / 波長: 0.9795 Å
/ ビームライン: 32-ID / 波長: 0.9795 Å 解析
解析 分子置換 / 解像度: 1.64→36.04 Å / σ(F): 0 /
分子置換 / 解像度: 1.64→36.04 Å / σ(F): 0 /  ムービー
ムービー コントローラー
コントローラー














 PDBj
PDBj






