+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1usq | ||||||
|---|---|---|---|---|---|---|---|
| Title | Complex of E. Coli DraE adhesin with Chloramphenicol | ||||||
 Components Components | DR HEMAGGLUTININ STRUCTURAL SUBUNIT | ||||||
 Keywords Keywords | ADHESIN / DRAE / FIMBRIAL ADHESIN / CHLORAMPHENICOL / UPEC / DAEC | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Anderson, K.L. / Billington, J. / Pettigrew, D. / Cota, E. / Roversi, P. / Simpson, P. / Chen, H.A. / Urvil, P. / Dumerle, L. / Barlow, P. ...Anderson, K.L. / Billington, J. / Pettigrew, D. / Cota, E. / Roversi, P. / Simpson, P. / Chen, H.A. / Urvil, P. / Dumerle, L. / Barlow, P. / Medof, E. / Smith, R.A.G. / Nowicki, B. / Le Bouguenec, C. / Lea, S.M. / Matthews, S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol Authors: Pettigrew, D. / Anderson, K.L. / Billington, J. / Cota, E. / Simpson, P. / Urvil, P. / Rabuzin, F. / Roversi, P. / Nowicki, B. / Du Merle, L. / Le Bouguenec, C. / Matthews, S. / Lea, S.M. #1:  Journal: Mol.Cell / Year: 2004 Journal: Mol.Cell / Year: 2004Title: An Atomic Resolution Model for Assmebly, Architecture,and Function of the Dr Adhesins Authors: Anderson, K.L. / Billington, J. / Pettigrew, D. / Cota, E. / Simpson, P. / Roversi, P. / Chen, H.A. / Urvil, P. / Du Merle, L. / Barlow, P.N. / Medof, M.E. / Smith, R.A.G. / Nowicki, B. / Le ...Authors: Anderson, K.L. / Billington, J. / Pettigrew, D. / Cota, E. / Simpson, P. / Roversi, P. / Chen, H.A. / Urvil, P. / Du Merle, L. / Barlow, P.N. / Medof, M.E. / Smith, R.A.G. / Nowicki, B. / Le Bouguenec, C. / Lea, S.M. / Matthews, S. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1usq.cif.gz 1usq.cif.gz | 188.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1usq.ent.gz pdb1usq.ent.gz | 151.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1usq.json.gz 1usq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/us/1usq https://data.pdbj.org/pub/pdb/validation_reports/us/1usq ftp://data.pdbj.org/pub/pdb/validation_reports/us/1usq ftp://data.pdbj.org/pub/pdb/validation_reports/us/1usq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1uszSC  1ut1C  1ut2C C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||
| 3 | 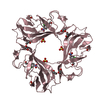
| ||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
| ||||||||||||||||||||||||
| Details | THERE ARE SIX TRIMERS IN THE CRYSTAL CELL , EACH LINKEDINTERNALLY BY 3 INTERMOLECULAR DISULPHIDE BONDS OF THEMONOMER WITH TWO OF ITS THREEFOLD-RELATED COPIES |
- Components
Components
| #1: Protein | Mass: 16169.844 Da / Num. of mol.: 6 / Mutation: YES Source method: isolated from a genetically manipulated source Details: COMPLEX WITH CHLORAMPHENICOL / Source: (gene. exp.)   #2: Chemical | ChemComp-SO4 / #3: Chemical | ChemComp-EDO / #4: Chemical | ChemComp-CLM / #5: Water | ChemComp-HOH / | Compound details | ENGINEERED MUTATION IN CHAINS A-F, GLY 21 TO ALA 21 ENGINEERED MUTATION IN CHAINS A-F, SER 22 TO ...ENGINEERED | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 50 % / Description: 20% EGL AS CRYOPROTECTANT |
|---|---|
| Crystal grow | pH: 7 Details: 1.7 M AMMONIUM SULPHATE 0.1M TRIS-HCL PH 7.0, 2.8 MM CHLORAMPHENICOL |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX9.6 / Wavelength: 0.87 / Beamline: PX9.6 / Wavelength: 0.87 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Oct 15, 2003 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.87 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→15 Å / Num. obs: 71543 / % possible obs: 99.8 % / Redundancy: 5.8 % / Biso Wilson estimate: 0.65 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 4.2 |
| Reflection shell | Resolution: 1.9→2 Å / Redundancy: 5.8 % / Rmerge(I) obs: 0.33 / Mean I/σ(I) obs: 1.4 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1USZ Resolution: 1.9→15 Å / Isotropic thermal model: TNT BCORREL / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: TNT CSDX_PROTGEO / Details: MAXIMUM LIKELIHOOD BUSTER-TNT REFINEMENT.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: BABINET SCALING / Bsol: 55 Å2 / ksol: 0.47 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→15 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj