[English] 日本語
 Yorodumi
Yorodumi- PDB-1sem: STRUCTURAL DETERMINANTS OF PEPTIDE-BINDING ORIENTATION AND OF SEQ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1sem | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURAL DETERMINANTS OF PEPTIDE-BINDING ORIENTATION AND OF SEQUENCE SPECIFICITY IN SH3 DOMAINS | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNAL TRANSDUCTION PROTEIN / SRC-HOMOLOGY 3 (SH3) DOMAIN / PEPTIDE-BINDING PROTEIN / GUANINE NUCLEOTIDE EXCHANGE FACTOR | ||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by SCF-KIT / Regulation of KIT signaling / GRB2 events in ERBB2 signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / NCAM signaling for neurite out-growth / : / PI-3K cascade:FGFR1 / FRS-mediated FGFR1 signaling / SHC-mediated cascade:FGFR2 / FRS-mediated FGFR2 signaling ...Signaling by SCF-KIT / Regulation of KIT signaling / GRB2 events in ERBB2 signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / NCAM signaling for neurite out-growth / : / PI-3K cascade:FGFR1 / FRS-mediated FGFR1 signaling / SHC-mediated cascade:FGFR2 / FRS-mediated FGFR2 signaling / SHC-mediated cascade:FGFR3 / FRS-mediated FGFR3 signaling / FRS-mediated FGFR4 signaling / : / PI-3K cascade:FGFR4 / Insulin receptor signalling cascade / MET activates RAS signaling / : / RHOU GTPase cycle / FLT3 Signaling / PIP3 activates AKT signaling / GRB2 events in EGFR signaling / SHC1 events in EGFR signaling / PI3K events in ERBB2 signaling / Regulation of actin dynamics for phagocytic cup formation / EGFR Transactivation by Gastrin / RHO GTPases Activate WASPs and WAVEs / RAF/MAP kinase cascade / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / Downstream signal transduction / : / : / regulation of nematode larval development / Negative regulation of MET activity / EGFR downregulation / Cargo recognition for clathrin-mediated endocytosis / Regulation of signaling by CBL / regulation of vulval development / Clathrin-mediated endocytosis / regulation of cell projection organization / male genitalia development / COP9 signalosome / epidermal growth factor receptor binding / muscle organ development / regulation of MAPK cascade / phosphotyrosine residue binding / regulation of cell migration / epidermal growth factor receptor signaling pathway / signal transduction / nucleoplasm / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Lim, W.A. / Richards, F.M. / Fox, R.O. | ||||||
 Citation Citation |  Journal: Nature / Year: 1994 Journal: Nature / Year: 1994Title: Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Authors: Lim, W.A. / Richards, F.M. / Fox, R.O. #1:  Journal: Nature / Year: 1993 Journal: Nature / Year: 1993Title: The Sh2 and SH3 Domains of Mammalian Grb2 Couple the Egf Receptor to the Ras Activator Msos1 Authors: Rozakis-Adcock, M. / Fernley, R. / Wade, J. / Pawson, T. / Bowtell, D. #2:  Journal: Nature / Year: 1992 Journal: Nature / Year: 1992Title: C. Elegans Cell-Signalling Gene Sem-5 Encodes a Protein with Sh2 and SH3 Domains Authors: Clark, S.G. / Stern, M.J. / Horvitz, H.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1sem.cif.gz 1sem.cif.gz | 39.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1sem.ent.gz pdb1sem.ent.gz | 27.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1sem.json.gz 1sem.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/se/1sem https://data.pdbj.org/pub/pdb/validation_reports/se/1sem ftp://data.pdbj.org/pub/pdb/validation_reports/se/1sem ftp://data.pdbj.org/pub/pdb/validation_reports/se/1sem | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 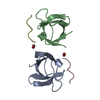
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6799.510 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein/peptide | Mass: 1100.339 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #3: Water | ChemComp-HOH / | Has protein modification | Y | Source details | MOLECULE_NAME: MSOS, 10-RESIDUE PEPTIDE (ACE-PRO-PRO-PRO-VAL-PRO-PRO-ARG-ARG-ARG). PEPTIDE IS FROM ...MOLECULE_NAME: MSOS, 10-RESIDUE PEPTIDE (ACE-PRO-PRO-PRO-VAL-PRO-PRO-ARG-ARG-ARG). PEPTIDE IS FROM BIOLOGICAL | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.03 Å3/Da / Density % sol: 39.46 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 8 / Method: vapor diffusion | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: MACSCIENCE / Detector: IMAGE PLATE / Date: Jul 24, 1994 |
| Radiation | Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Num. obs: 6912 / % possible obs: 92.5 % / Observed criterion σ(I): 2 / Redundancy: 3 % / Rmerge(I) obs: 0.08 |
| Reflection | *PLUS Highest resolution: 2 Å / Rmerge(I) obs: 0.08 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→7 Å / σ(F): 2 Details: SSBOND CRYSTALS GROWN IN THE PRESENCE OF DTT; DISULFIDE BOND THOUGHT TO FORM AFTER INITIAL EXPOSURE TO X-RAYS; CAUSED STABLE SHIFT IN UNIT CELL DIMENSIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller











 PDBj
PDBj
