+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1s34 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
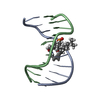
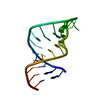
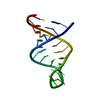
| Title | Solution structure of residues 907-929 from Rous Sarcoma Virus | ||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / stem-loop / tetraloop / bulge | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / Simulated annealing. Refinement using torsion angle dynamics, the DELPHIC database |  Authors AuthorsCabello-Villegas, J. / Giles, K.E. / Soto, A.M. / Yu, P. / Beemon, K.L. / Wang, Y.X. |  Citation Citation Journal: Rna / Year: 2004 Journal: Rna / Year: 2004Title: Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor. Authors: Cabello-Villegas, J. / Giles, K.E. / Soto, A.M. / Yu, P. / Mougin, A. / Beemon, K.L. / Wang, Y.X. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1s34.cif.gz 1s34.cif.gz | 213.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1s34.ent.gz pdb1s34.ent.gz | 179.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1s34.json.gz 1s34.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s3/1s34 https://data.pdbj.org/pub/pdb/validation_reports/s3/1s34 ftp://data.pdbj.org/pub/pdb/validation_reports/s3/1s34 ftp://data.pdbj.org/pub/pdb/validation_reports/s3/1s34 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 7350.335 Da / Num. of mol.: 1 / Fragment: Pseudo 5'-splice site / Source method: obtained synthetically / Details: In vitro transcription |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: RDC restraints were obtained for the base one bond H-C and H-N using HSQC experiments. RDC restraints for the ribose were obtained from measurements on an Relay HCCH-COSY (Vallurupalli and ...Text: RDC restraints were obtained for the base one bond H-C and H-N using HSQC experiments. RDC restraints for the ribose were obtained from measurements on an Relay HCCH-COSY (Vallurupalli and Moore, J Biomol NMR. 2002. 24(1):63-6). Anisotropic conditions were ~20 mg/ml pf1 phage. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Simulated annealing. Refinement using torsion angle dynamics, the DELPHIC database Software ordinal: 1 Details: DELPHIC torsion angle potential was used for the whole molecule. The base position potential was excluded for residues 913, and 917 to 920. Protocol from Clore and Kuszewski (J Am Chem Soc. 2003. 125(6):1518-25) | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 15 |
 Movie
Movie Controller
Controller









 PDBj
PDBj





























 NMRPipe
NMRPipe