+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rrr | ||||||
|---|---|---|---|---|---|---|---|
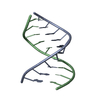
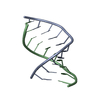
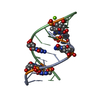
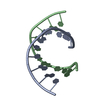
| Title | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA / RNA DUPLEX / PURINE/PYRIMIDINE-RICH STRANDS / A-FORM | ||||||
| Function / homology | RNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Gyi, J.I. / Lane, A.N. / Conn, G.L. / Brown, T. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Authors: Gyi, J.I. / Lane, A.N. / Conn, G.L. / Brown, T. #1:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Comparison of the Thermodynamic Stabilities and Solution Conformations of DNA.RNA Hybrids Containing Purine-Rich and Pyrimidine-Rich Strands with DNA and RNA Duplexes Authors: Gyi, J.I. / Conn, G.L. / Lane, A.N. / Brown, T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rrr.cif.gz 1rrr.cif.gz | 126 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rrr.ent.gz pdb1rrr.ent.gz | 105.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rrr.json.gz 1rrr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rr/1rrr https://data.pdbj.org/pub/pdb/validation_reports/rr/1rrr ftp://data.pdbj.org/pub/pdb/validation_reports/rr/1rrr ftp://data.pdbj.org/pub/pdb/validation_reports/rr/1rrr | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 3287.076 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: RNA chain | Mass: 3051.804 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 7 / Temperature: 303 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| Software | Name:  AMBER / Classification: refinement AMBER / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||||||||||||||
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: DISTANCE CONSTRAINTS FOR GLYCOSIDIC TORSION ANGLES, RIBOSE PSEUDOROTATIONAL PHASE ANGLES AND AMPLITUDES CALCULATED FROM NOE BUILD-UP CURVES AND COUPLING CONSTANTS BY A LEAST SQUARES/GRID- ...Details: DISTANCE CONSTRAINTS FOR GLYCOSIDIC TORSION ANGLES, RIBOSE PSEUDOROTATIONAL PHASE ANGLES AND AMPLITUDES CALCULATED FROM NOE BUILD-UP CURVES AND COUPLING CONSTANTS BY A LEAST SQUARES/GRID-SEARCH METHOD IMPLEMENTED IN NUCFIT AND PFIT (A.N. LANE, NIMR, UK). INTERNUCLEOTIDE DISTANCE CONSTRAINTS CALCULATED FROM NOE BUILD-UP CURVES. STRUCTURES CALCULATED BY SIMULATED ANNEALING/RMD PROTOCOL WITHIN DISCOVER 95.0 USING AN AMBER FORCEFIELD AND A DISTANCE DEPENDENT DIELECTRIC CONSTANT. | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST POTENTIAL ENERGY AND MINIMAL VIOLATIONS AND ACCEPTABLE STEREOCHEMISTRY Conformers calculated total number: 32 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller









 PDBj
PDBj