[English] 日本語
 Yorodumi
Yorodumi- PDB-1rl2: RIBOSOMAL PROTEIN L2 RNA-BINDING DOMAIN FROM BACILLUS STEAROTHERM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rl2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | RIBOSOMAL PROTEIN L2 RNA-BINDING DOMAIN FROM BACILLUS STEAROTHERMOPHILUS | ||||||
 Components Components | PROTEIN (RIBOSOMAL PROTEIN L2) | ||||||
 Keywords Keywords | RIBOSOMAL PROTEIN / RNA-BINDING DOMAIN / PEPTIDYLTRANSFEREASE CENTER | ||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / cytoplasmic translation / rRNA binding / structural constituent of ribosome Similarity search - Function | ||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.3 Å MAD / Resolution: 2.3 Å | ||||||
 Authors Authors | Nakagawa, A. / Hosaka, H. / Nakashima, T. / Tanaka, I. | ||||||
 Citation Citation |  Journal: EMBO J. / Year: 1999 Journal: EMBO J. / Year: 1999Title: The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome. Authors: Nakagawa, A. / Nakashima, T. / Taniguchi, M. / Hosaka, H. / Kimura, M. / Tanaka, I. #1:  Journal: J.Struct.Biol. / Year: 1998 Journal: J.Struct.Biol. / Year: 1998Title: Crystallization and preliminary X-ray crystallographic study of a 23S rRNA binding domain of the ribosomal protein L2 from Bacillus stearothermophilus Authors: Nakashima, T. / Kimura, M. / Nakagawa, A. / Tanaka, I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rl2.cif.gz 1rl2.cif.gz | 65.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rl2.ent.gz pdb1rl2.ent.gz | 48.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rl2.json.gz 1rl2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rl/1rl2 https://data.pdbj.org/pub/pdb/validation_reports/rl/1rl2 ftp://data.pdbj.org/pub/pdb/validation_reports/rl/1rl2 ftp://data.pdbj.org/pub/pdb/validation_reports/rl/1rl2 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 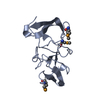
| |||||||||||||||
| 2 | 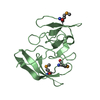
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS oper: (Code: given Matrix: (-0.999427, -0.006932, 0.033133), Vector: |
- Components
Components
| #1: Protein | Mass: 14930.819 Da / Num. of mol.: 2 / Mutation: METHIONINES ARE SUBSTITUED BY SELONOMETHIONINE Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria)Plasmid: PET-22B / Gene (production host): L2 / Production host:  #2: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43 % Description: DATA WERE COLLECTED USING THE WEISSENBERG METHOD | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6.5 Details: PROTEIN WAS CRYSTALLIZED FROM 15% PEK20000 IN 0.1M MES PH6, pH 6.5 | ||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / Method: vapor diffusion, hanging dropDetails: drop consists of equal volume of protein and reservoir solutions | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: BL-18B / Wavelength: 0.9000, 0.9785, 0.9788 / Beamline: BL-18B / Wavelength: 0.9000, 0.9785, 0.9788 | ||||||||||||
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: May 15, 1998 / Details: FOCUSING MIRRORS | ||||||||||||
| Radiation | Monochromator: SI(111) / Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength |
| ||||||||||||
| Reflection | Resolution: 2.3→40 Å / Num. obs: 11430 / % possible obs: 98.6 % / Redundancy: 3.9 % / Biso Wilson estimate: 15 Å2 / Rmerge(I) obs: 0.084 / Net I/σ(I): 8.3 | ||||||||||||
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 3.9 % / Rmerge(I) obs: 0.354 / Mean I/σ(I) obs: 2.1 / % possible all: 96.8 | ||||||||||||
| Reflection | *PLUS Num. measured all: 45101 | ||||||||||||
| Reflection shell | *PLUS % possible obs: 96.8 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.3→20 Å / Rfactor Rfree error: 0.008 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 MAD / Resolution: 2.3→20 Å / Rfactor Rfree error: 0.008 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 46.4 Å2 / ksol: 0.36 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 29.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Refine-ID: X-RAY DIFFRACTION / Weight Biso : 0.2 / Weight position: 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.44 Å / Rfactor Rfree error: 0.024 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.5 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.203 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.258 |
 Movie
Movie Controller
Controller











 PDBj
PDBj
