+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rju | ||||||
|---|---|---|---|---|---|---|---|
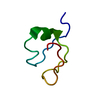
| Title | Crystal structure of a truncated form of yeast copper thionein | ||||||
 Components Components | Metallothionein | ||||||
 Keywords Keywords | METAL BINDING PROTEIN / Yeast Cu(I) metallothionein / Cu(I) metallothionein fragments / Cu(I)-thiolate | ||||||
| Function / homology |  Function and homology information Function and homology informationdetoxification of cadmium ion / detoxification of copper ion / response to copper ion / superoxide dismutase activity / antioxidant activity / cadmium ion binding / removal of superoxide radicals / mitochondrial intermembrane space / copper ion binding / cytosol Similarity search - Function | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.44 Å SAD / Resolution: 1.44 Å | ||||||
 Authors Authors | Calderone, V. / Dolderer, B. / Hartmann, H.J. / Echner, H. / Luchinat, C. / Del Bianco, C. / Mangani, S. / Weser, U. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2005 Journal: Proc.Natl.Acad.Sci.USA / Year: 2005Title: The Crystal Structure of Yeast Copper Thionein: the solution of a long lasting Enigma. Authors: Calderone, V. / Dolderer, B. / Hartmann, H.J. / Echner, H. / Luchinat, C. / Del Bianco, C. / Mangani, S. / Weser, U. #1:  Journal: Eur.J.Biochem. / Year: 2000 Journal: Eur.J.Biochem. / Year: 2000Title: High resolution solution structure of the protein part of Cu7 metallothionein Authors: Bertini, I. / Hartmann, H.J. / Klein, T. / Liu, G. / Luchinat, C. / Weser, U. #2:  Journal: J.Biol.Inorg.Chem. / Year: 2003 Journal: J.Biol.Inorg.Chem. / Year: 2003Title: The Cu(I)(7) cluster in yeast copper thionein survives major shortening of the polypeptide backbone as deduced from electronic absorption, circular dichroism, luminescence and (1)H NMR Authors: Luchinat, C. / Dolderer, B. / Del Bianco, C. / Echner, H. / Hartmann, H.J. / Voelter, W. / Weser, U. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rju.cif.gz 1rju.cif.gz | 26.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rju.ent.gz pdb1rju.ent.gz | 19.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rju.json.gz 1rju.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1rju_validation.pdf.gz 1rju_validation.pdf.gz | 410 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1rju_full_validation.pdf.gz 1rju_full_validation.pdf.gz | 410 KB | Display | |
| Data in XML |  1rju_validation.xml.gz 1rju_validation.xml.gz | 4 KB | Display | |
| Data in CIF |  1rju_validation.cif.gz 1rju_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rj/1rju https://data.pdbj.org/pub/pdb/validation_reports/rj/1rju ftp://data.pdbj.org/pub/pdb/validation_reports/rj/1rju ftp://data.pdbj.org/pub/pdb/validation_reports/rj/1rju | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein/peptide | Mass: 3847.259 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: The protein was Chemically synthesized. The sequence of the protein IS NATURALLY FOUND IN Saccharomyces cerevisiae (Baker's yeast). References: UniProt: P07215, UniProt: P0CX80*PLUS | ||
|---|---|---|---|
| #2: Chemical | ChemComp-CU1 / #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52.73 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7.2 Details: 250 mM KH2PO4, 1mM LiBr, pH 7.2, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: BW7A / Wavelength: 0.9192 Å / Beamline: BW7A / Wavelength: 0.9192 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Nov 5, 2003 |
| Radiation | Monochromator: Double crystal focusing monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9192 Å / Relative weight: 1 |
| Reflection | Resolution: 1.44→27.84 Å / Num. all: 7922 / Num. obs: 7922 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 21.8 % / Biso Wilson estimate: 9.155 Å2 / Rmerge(I) obs: 0.07 / Rsym value: 0.07 / Net I/σ(I): 9.3 |
| Reflection shell | Resolution: 1.44→1.52 Å / Redundancy: 22.4 % / Rmerge(I) obs: 0.358 / Mean I/σ(I) obs: 2.1 / Num. unique all: 1117 / Rsym value: 0.358 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.44→27.84 Å / Cor.coef. Fo:Fc: 0.955 / Cor.coef. Fo:Fc free: 0.944 / SU B: 0.568 / SU ML: 0.024 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.056 / ESU R Free: 0.052 / Stereochemistry target values: MAXIMUM LIKELIHOOD SAD / Resolution: 1.44→27.84 Å / Cor.coef. Fo:Fc: 0.955 / Cor.coef. Fo:Fc free: 0.944 / SU B: 0.568 / SU ML: 0.024 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.056 / ESU R Free: 0.052 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. SOME EXTRA ELECTRON DENSITY CORRESPONDING TO 7 ATOMS HAS BEEN LEFT UNINTERPRETED SINCE NEITHER WATER MOLECULES NOR OTHER MOLECULES PRESENT ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. SOME EXTRA ELECTRON DENSITY CORRESPONDING TO 7 ATOMS HAS BEEN LEFT UNINTERPRETED SINCE NEITHER WATER MOLECULES NOR OTHER MOLECULES PRESENT IN THE CRYSTALLISATION SOLUTION COULD ACCOUNT FOR IT.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 12.889 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.44→27.84 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.44→1.478 Å / Total num. of bins used: 20 /
|
 Movie
Movie Controller
Controller











 PDBj
PDBj


