+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 1r8e | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | Crystal Structure of BmrR Bound to DNA at 2.4A Resolution | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード | TRANSCRIPTION/DNA / PROTEIN-DNA COMPLEX / MERR-FAMILY TRANSCRIPTION ACTIVATOR / MULTIDRUG-BINDING PROTEIN / TRANSCRIPTION-DNA COMPLEX | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報DNA-binding transcription factor activity / regulation of DNA-templated transcription / DNA binding 類似検索 - 分子機能 | ||||||
| 生物種 |  | ||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.4 Å 分子置換 / 解像度: 2.4 Å | ||||||
 データ登録者 データ登録者 | Newberry, K.J. / Brennan, R.G. | ||||||
 引用 引用 |  ジャーナル: J.Biol.Chem. / 年: 2004 ジャーナル: J.Biol.Chem. / 年: 2004タイトル: The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. 著者: Newberry, K.J. / Brennan, R.G. #1:  ジャーナル: Nature / 年: 2001 ジャーナル: Nature / 年: 2001タイトル: Crystal structure of the transcription activator BmrR bound to DNA and a drug 著者: Heldwein, E.E. / Brennan, R.G. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  1r8e.cif.gz 1r8e.cif.gz | 92.2 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb1r8e.ent.gz pdb1r8e.ent.gz | 66.4 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  1r8e.json.gz 1r8e.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/r8/1r8e https://data.pdbj.org/pub/pdb/validation_reports/r8/1r8e ftp://data.pdbj.org/pub/pdb/validation_reports/r8/1r8e ftp://data.pdbj.org/pub/pdb/validation_reports/r8/1r8e | HTTPS FTP |
|---|
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 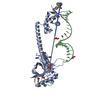
| ||||||||
| 単位格子 |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| 詳細 | The second part of the biological assembly is generate by the operations: x, -y, 1/2 - z |
- 要素
要素
-DNA鎖 / タンパク質 , 2種, 2分子 BA
| #1: DNA鎖 | 分子量: 7058.536 Da / 分子数: 1 / 由来タイプ: 合成 |
|---|---|
| #2: タンパク質 | 分子量: 32621.281 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)   |
-非ポリマー , 4種, 148分子 





| #3: 化合物 | ChemComp-GOL / #4: 化合物 | ChemComp-P4P / | #5: 化合物 | ChemComp-IMD / #6: 水 | ChemComp-HOH / | |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 5.29 Å3/Da / 溶媒含有率: 76.55 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 結晶化 | 温度: 273 K / 手法: 蒸気拡散法, ハンギングドロップ法 / pH: 6.5 詳細: Imidazole, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 273K | ||||||||||||||||||||
| 溶液の組成 |
|
-データ収集
| 回折 | 平均測定温度: 100 K |
|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  SSRL SSRL  / ビームライン: BL11-1 / 波長: 0.9537 Å / ビームライン: BL11-1 / 波長: 0.9537 Å |
| 検出器 | タイプ: ADSC QUANTUM 4 / 検出器: CCD / 日付: 2002年12月22日 / 詳細: single crystal monochromator |
| 放射 | モノクロメーター: Si / プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray |
| 放射波長 | 波長: 0.9537 Å / 相対比: 1 |
| 反射 | 解像度: 2.4→86.39 Å / Num. all: 33967 / Num. obs: 33613 / % possible obs: 99 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Biso Wilson estimate: 50.9 Å2 |
| 反射 シェル | 解像度: 2.4→2.55 Å / % possible all: 99.9 |
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 分子置換開始モデル: PDB ENTRY 1EXJ 解像度: 2.4→86.39 Å / Rfactor Rfree error: 0.006 / Isotropic thermal model: RESTRAINED / 交差検証法: THROUGHOUT / σ(F): 0 / 立体化学のターゲット値: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 溶媒モデル: FLAT MODEL / Bsol: 57.6217 Å2 / ksol: 0.349872 e/Å3 | ||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso mean: 60.2 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 解像度: 2.4→86.39 Å
| ||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||
| LS精密化 シェル | 解像度: 2.4→2.55 Å / Rfactor Rfree error: 0.02 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj














































