[English] 日本語
 Yorodumi
Yorodumi- PDB-1r3x: INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1r3x | ||||||
|---|---|---|---|---|---|---|---|
| Title | INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA-RNA HYBRID / OLIGONUCLEOTIDE / TRIPLEX / RNA THIRD STRAND / DNA-RNA COMPLEX | ||||||
| Function / homology | DNA / RNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, SIMULATED ANNEALING | ||||||
 Authors Authors | Gotfredsen, C.H. / Schultze, P. / Feigon, J. | ||||||
 Citation Citation | Journal: J.Am.Chem.Soc. / Year: 1998 Title: Solution Structure of an Intramolecular Pyrimidine-Purine-Pyrimidine Triplex Containing an RNA Third Strand Authors: Gotfredsen, C.H. / Schultze, P. / Feigon, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1r3x.cif.gz 1r3x.cif.gz | 161.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1r3x.ent.gz pdb1r3x.ent.gz | 123.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1r3x.json.gz 1r3x.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1r3x_validation.pdf.gz 1r3x_validation.pdf.gz | 337.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1r3x_full_validation.pdf.gz 1r3x_full_validation.pdf.gz | 498.1 KB | Display | |
| Data in XML |  1r3x_validation.xml.gz 1r3x_validation.xml.gz | 28.9 KB | Display | |
| Data in CIF |  1r3x_validation.cif.gz 1r3x_validation.cif.gz | 40.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r3/1r3x https://data.pdbj.org/pub/pdb/validation_reports/r3/1r3x ftp://data.pdbj.org/pub/pdb/validation_reports/r3/1r3x ftp://data.pdbj.org/pub/pdb/validation_reports/r3/1r3x | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 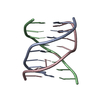
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2508.694 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: HEXAKIS (ETHYLENE GLYCOL) LINKERS BETWEEN A 8 AND T 9 AND BETWEEN T 16 AND U 17. C 18, C 20, AND C 22 ARE PROTONATED AT N3 |
|---|---|
| #2: DNA chain | Mass: 2343.552 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: HEXAKIS (ETHYLENE GLYCOL) LINKERS BETWEEN A 8 AND T 9 AND BETWEEN T 16 AND U 17. C 18, C 20, AND C 22 ARE PROTONATED AT N3 |
| #3: RNA chain | Mass: 2401.417 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: HEXAKIS (ETHYLENE GLYCOL) LINKERS BETWEEN A 8 AND T 9 AND BETWEEN T 16 AND U 17. C 18, C 20, AND C 22 ARE PROTONATED AT N3 |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 5.7 / Temperature: 303 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: DISTANCE GEOMETRY, SIMULATED ANNEALING / Software ordinal: 1 Details: LAST REFINEMENT STEP USED RELAXATION MATRIX REFINEMENT. | ||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST OVERALL ENERGY / Conformers calculated total number: 20 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller











 PDBj
PDBj HSQC
HSQC