[English] 日本語
 Yorodumi
Yorodumi- PDB-1qwb: NMR structure of 5'-r(GGACACGAAAUCCCGAAGUAGUGUCC)-3' : an RNA hai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qwb | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of 5'-r(GGACACGAAAUCCCGAAGUAGUGUCC)-3' : an RNA hairpin containing the in vitro selected consensus sequence for nucleolin RBD12 | ||||||
 Components Components | sNRE26 | ||||||
 Keywords Keywords | RNA / A-form helix / loop E motif / S-turn / disordered hairpin loop | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Finger, L.D. / Trantirek, L. / Johansson, C. / Feigon, J. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2003 Journal: Nucleic Acids Res. / Year: 2003Title: Solution Structures of Stem-loop RNAs that Bind the Two N-terminal RNA-binding Domains of Nucleolin Authors: Finger, L.D. / Trantirek, L. / Johansson, C. / Feigon, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qwb.cif.gz 1qwb.cif.gz | 277.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qwb.ent.gz pdb1qwb.ent.gz | 232.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qwb.json.gz 1qwb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1qwb_validation.pdf.gz 1qwb_validation.pdf.gz | 322.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1qwb_full_validation.pdf.gz 1qwb_full_validation.pdf.gz | 431.5 KB | Display | |
| Data in XML |  1qwb_validation.xml.gz 1qwb_validation.xml.gz | 15.6 KB | Display | |
| Data in CIF |  1qwb_validation.cif.gz 1qwb_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qw/1qwb https://data.pdbj.org/pub/pdb/validation_reports/qw/1qwb ftp://data.pdbj.org/pub/pdb/validation_reports/qw/1qwb ftp://data.pdbj.org/pub/pdb/validation_reports/qw/1qwb | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 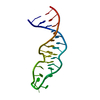
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8366.066 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: RNA WAS PREPARED BY In vitro transcription with T7 RNA polymerase |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: Assignment methodology for this molecule is described in J. Biomol. NMR 9, 259-272 (1997) and Prog. in Nuc. Magn. Resonan. Spec. 32, 287-387 (1998). |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted,structures with acceptable covalent geometry,structures with favorable non-bond energy,structures with the least restraint ...Conformer selection criteria: all calculated structures submitted,structures with acceptable covalent geometry,structures with favorable non-bond energy,structures with the least restraint violations,structures with the lowest energy Conformers calculated total number: 17 / Conformers submitted total number: 17 |
 Movie
Movie Controller
Controller










 PDBj
PDBj





























 HSQC
HSQC