[English] 日本語
 Yorodumi
Yorodumi- PDB-1pqt: REFINEMENT OF d(GCGAAGC) HAIRPIN STRUCTURE USING ONE- AND TWO-BON... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pqt | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | REFINEMENT OF d(GCGAAGC) HAIRPIN STRUCTURE USING ONE- AND TWO-BOND RESIDUAL DIPOLAR COUPLINGS | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / HAIRPIN / GA MISMATCH | Function / homology | DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / molecular dynamics |  Authors AuthorsPadrta, P. / Stefl, R. / Zidek, L. / Sklenar, V. |  Citation Citation Journal: J.Biomol.NMR / Year: 2002 Journal: J.Biomol.NMR / Year: 2002Title: Refinement of d(GCGAAGC) Hairpin Structure Using One- and Two-Bond Residual Dipolar Couplings Authors: Padrta, P. / Stefl, R. / Kralik, L. / Zidek, L. / Sklenar, V. History |
|
- Structure visualization
Structure visualization
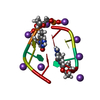
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pqt.cif.gz 1pqt.cif.gz | 57.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pqt.ent.gz pdb1pqt.ent.gz | 38.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pqt.json.gz 1pqt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1pqt_validation.pdf.gz 1pqt_validation.pdf.gz | 298.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1pqt_full_validation.pdf.gz 1pqt_full_validation.pdf.gz | 358.9 KB | Display | |
| Data in XML |  1pqt_validation.xml.gz 1pqt_validation.xml.gz | 3.2 KB | Display | |
| Data in CIF |  1pqt_validation.cif.gz 1pqt_validation.cif.gz | 4.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pq/1pqt https://data.pdbj.org/pub/pdb/validation_reports/pq/1pqt ftp://data.pdbj.org/pub/pdb/validation_reports/pq/1pqt ftp://data.pdbj.org/pub/pdb/validation_reports/pq/1pqt | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2147.437 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: THIS SEQUENCE OCCURS NATURALLY IN REPLICATION ORIGINS OF PHAGE PHIX174, IN HERPES SIMPLEX VIRUS, IN E. COLI HEAT-SHOCK GENE AND RRNA GENES. |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TRIPLE-RESONANCE NMR SPECTROSCOPY IN ISOTROPIC AND LIQUID CRYSTALLINE PHASE. |
- Sample preparation
Sample preparation
| Details | Contents: 6MM NATURAL ABUNDANCE D(GCGAAGC), 10MM PHOSPHATE BUFFER NA, D2O; 6MM NATURAL ABUNDANCE D(GCGAAGC), 10MM PHOSPHATE BUFFER NA, 90% H2O, 10% D2O; U-15N,13C, 0.5MM D(GCGAAGC), 10MM PHOSPHATE ...Contents: 6MM NATURAL ABUNDANCE D(GCGAAGC), 10MM PHOSPHATE BUFFER NA, D2O; 6MM NATURAL ABUNDANCE D(GCGAAGC), 10MM PHOSPHATE BUFFER NA, 90% H2O, 10% D2O; U-15N,13C, 0.5MM D(GCGAAGC), 10MM PHOSPHATE BUFFER NA, 90% H2O, 10% D2O |
|---|---|
| Sample conditions | Ionic strength: 10 mM / pH: 6.7 / Pressure: 1 atm / Temperature: 303 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 14 / Conformers submitted total number: 14 |
 Movie
Movie Controller
Controller







 PDBj
PDBj

 HSQC
HSQC