[English] 日本語
 Yorodumi
Yorodumi- PDB-1pcs: THE 2.15 A CRYSTAL STRUCTURE OF A TRIPLE MUTANT PLASTOCYANIN FROM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pcs | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE 2.15 A CRYSTAL STRUCTURE OF A TRIPLE MUTANT PLASTOCYANIN FROM THE CYANOBACTERIUM SYNECHOCYSTIS SP. PCC 6803 | ||||||
 Components Components | PLASTOCYANIN | ||||||
 Keywords Keywords | ELECTRON TRANSPORT / PLASTOCYANIN | ||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived thylakoid membrane / electron transfer activity / copper ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | ||||||
 Authors Authors | Romero, A. / De La Cerda, B. / Varela, P.F. / Navarro, J.A. / Hervas, M. / De La Rosa, M.A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: The 2.15 A crystal structure of a triple mutant plastocyanin from the cyanobacterium Synechocystis sp. PCC 6803. Authors: Romero, A. / De la Cerda, B. / Varela, P.F. / Navarro, J.A. / Hervas, M. / De la Rosa, M.A. #1:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Solution Structure of Reduced Plastocyanin from the Blue-Green Alga Anabaena Variabilis Authors: Badsberg, U. / Jorgensen, A.M. / Gesmar, H. / Led, J.J. / Hammerstad, J.M. / Jespersen, L.L. / Ulstrup, J. #2:  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Structure of Oxidized Poplar Plastocyanin at 1.6 A Resolution Authors: Guss, J.M. / Freeman, H.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pcs.cif.gz 1pcs.cif.gz | 38.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pcs.ent.gz pdb1pcs.ent.gz | 26.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pcs.json.gz 1pcs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1pcs_validation.pdf.gz 1pcs_validation.pdf.gz | 393.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1pcs_full_validation.pdf.gz 1pcs_full_validation.pdf.gz | 393.3 KB | Display | |
| Data in XML |  1pcs_validation.xml.gz 1pcs_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  1pcs_validation.cif.gz 1pcs_validation.cif.gz | 8.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pc/1pcs https://data.pdbj.org/pub/pdb/validation_reports/pc/1pcs ftp://data.pdbj.org/pub/pdb/validation_reports/pc/1pcs ftp://data.pdbj.org/pub/pdb/validation_reports/pc/1pcs | HTTPS FTP |
-Related structure data
| Related structure data |  1pcy S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 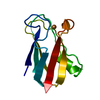
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10331.574 Da / Num. of mol.: 1 / Mutation: A42D, D47P, A63L Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Chemical | ChemComp-CU / |
| #3: Water | ChemComp-HOH / |
| Sequence details | WHEN SYNECHOCYSTIS PS IS ALIGNED WITH THE AMINO ACID SEQUENCE OF POPLAR PC, THE SEQUENCE OF ...WHEN SYNECHOCYS |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.94 Å3/Da / Density % sol: 37 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6 Details: PROTEIN WAS CRYSTALLIZED FROM 3.2M AMMONIUM SULFATE, 0.1M NA,K.PHOSPHATE, PH 6.0 WITH A PROTEIN CONCENTRATION OF 10 MG/ML | |||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 295 K |
|---|---|
| Diffraction source | Type: ENRAF-NONIUS / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Apr 1, 1996 |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.15→20.3 Å / Num. obs: 4422 / % possible obs: 92.7 % / Observed criterion σ(I): 2 / Redundancy: 5.5 % / Biso Wilson estimate: 12.72 Å2 / Rsym value: 0.065 / Net I/σ(I): 8 |
| Reflection shell | Resolution: 2.15→2.25 Å / Redundancy: 5.2 % / Mean I/σ(I) obs: 8.2 / Rsym value: 0.074 / % possible all: 98 |
| Reflection | *PLUS Num. measured all: 26699 / Rmerge(I) obs: 0.065 |
| Reflection shell | *PLUS % possible obs: 98 % / Rmerge(I) obs: 0.074 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1PCY  1pcy Resolution: 2.15→8 Å / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 11.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.19 Å / Luzzati d res low obs: 8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.15→2.3 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1F / Classification: refinement X-PLOR / Version: 3.1F / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Num. reflection obs: 4070 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.174 |
 Movie
Movie Controller
Controller









 PDBj
PDBj


