+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1p71 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Anabaena HU-DNA corcrystal structure (TR3) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA Binding Protein/DNA / protein-DNA complex / DNA bending / HU / DNA Binding Protein-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationheterocyst development / chromosome condensation / structural constituent of chromatin / DNA binding / cytosol Similarity search - Function | ||||||
| Biological species |  Anabaena sp. (bacteria) Anabaena sp. (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Swinger, K.S. / Lemberg, K.M. / Zhang, Y. / Rice, P.A. | ||||||
 Citation Citation |  Journal: Embo J. / Year: 2003 Journal: Embo J. / Year: 2003Title: Flexible DNA bending in HU-DNA cocrystal structures Authors: Swinger, K.S. / Lemberg, K.M. / Zhang, Y. / Rice, P.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1p71.cif.gz 1p71.cif.gz | 75.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1p71.ent.gz pdb1p71.ent.gz | 54.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1p71.json.gz 1p71.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1p71_validation.pdf.gz 1p71_validation.pdf.gz | 453.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1p71_full_validation.pdf.gz 1p71_full_validation.pdf.gz | 463.4 KB | Display | |
| Data in XML |  1p71_validation.xml.gz 1p71_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  1p71_validation.cif.gz 1p71_validation.cif.gz | 19.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p7/1p71 https://data.pdbj.org/pub/pdb/validation_reports/p7/1p71 ftp://data.pdbj.org/pub/pdb/validation_reports/p7/1p71 ftp://data.pdbj.org/pub/pdb/validation_reports/p7/1p71 | HTTPS FTP |
-Related structure data
| Related structure data |  1p51C  1p78C  1b8zS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 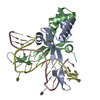
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The assymetric unit contains one functional complex composed of a protein homodimer and duplex DNA. |
- Components
Components
| #1: DNA chain | Mass: 6379.131 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: chemically synthesized DNA #2: Protein | Mass: 10089.584 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Anabaena sp. (bacteria) / Gene: HUP OR HANA OR ASR3935 / Plasmid: pET21a (pETAHU) / Production host: Anabaena sp. (bacteria) / Gene: HUP OR HANA OR ASR3935 / Plasmid: pET21a (pETAHU) / Production host:  #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.61 Å3/Da / Density % sol: 52.45 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: Peg 5000 monomethyl ether, glycerol, tris, jeffamine, potassium chloride, calcium chloride, sodium azide, pH 7.5, VAPOR DIFFUSION, HANGING DROP, temperature 292K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 14-BM-C / Wavelength: 1 Å / Beamline: 14-BM-C / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Feb 11, 2001 |
| Radiation | Monochromator: Ge 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→100 Å / Num. all: 20602 / Num. obs: 20602 / % possible obs: 72.4 % / Observed criterion σ(I): -3 / Redundancy: 7.6 % / Rmerge(I) obs: 0.041 |
| Reflection shell | Resolution: 1.9→2.01 Å / Redundancy: 1.6 % / Rmerge(I) obs: 0.3 / % possible all: 20.9 |
| Reflection | *PLUS Lowest resolution: 100 Å / Rmerge(I) obs: 0.04 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1B8Z with nonidentical sidechains pruned back to a common atom Resolution: 1.9→25 Å / Cor.coef. Fo:Fc: 0.929 / Cor.coef. Fo:Fc free: 0.905 / SU B: 7.985 / SU ML: 0.21 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.27 / ESU R Free: 0.224 Details: Data are anisotropic with limits 1.9 X 2.5 x 2.0. Data were truncated to an ellipsoid and reflections with an average (I/sigI) ratio less than 2 were removed. The completeness above is ...Details: Data are anisotropic with limits 1.9 X 2.5 x 2.0. Data were truncated to an ellipsoid and reflections with an average (I/sigI) ratio less than 2 were removed. The completeness above is underestimated. When truncation is factored in, data in refinement are 91% complete. The following residues in chain A and B have some sidechain atoms with 0.00 occupancy: A3, A12, A13, A18, A19, A34, A45, A59, B3, B12, B18, B59, B67, B83, B84.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.952 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→25 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→2.002 Å / Total num. of bins used: 10 /
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.9 Å / Lowest resolution: 25 Å / % reflection Rfree: 5 % / Rfactor Rfree: 0.288 / Rfactor Rwork: 0.244 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller













 PDBj
PDBj







































