[English] 日本語
 Yorodumi
Yorodumi- PDB-1p2p: STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 AT 2.6 ANGSTROMS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1p2p | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 AT 2.6 ANGSTROMS RESOLUTION AND COMPARISON WITH BOVINE PHOSPHOLIPASE A2 | ||||||
 Components Components | PHOSPHOLIPASE A2 | ||||||
 Keywords Keywords | CARBOXYLIC ESTER HYDROLASE | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of D-glucose import across plasma membrane / positive regulation of podocyte apoptotic process / phosphatidylglycerol metabolic process / A2-type glycerophospholipase activity / phosphatidylcholine metabolic process / leukotriene biosynthetic process / neutrophil mediated immunity / bile acid binding / phospholipase A2 / : ...regulation of D-glucose import across plasma membrane / positive regulation of podocyte apoptotic process / phosphatidylglycerol metabolic process / A2-type glycerophospholipase activity / phosphatidylcholine metabolic process / leukotriene biosynthetic process / neutrophil mediated immunity / bile acid binding / phospholipase A2 / : / positive regulation of calcium ion transport into cytosol / lipid catabolic process / neutrophil chemotaxis / positive regulation of interleukin-8 production / phospholipid binding / cellular response to insulin stimulus / positive regulation of fibroblast proliferation / positive regulation of immune response / fatty acid biosynthetic process / positive regulation of MAPK cascade / intracellular signal transduction / signaling receptor binding / positive regulation of cell population proliferation / calcium ion binding / cell surface / positive regulation of transcription by RNA polymerase II / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.6 Å X-RAY DIFFRACTION / Resolution: 2.6 Å | ||||||
 Authors Authors | Dijkstra, B.W. / Renetseder, R. / Kalk, K.H. / Hol, W.G.J. / Drenth, J. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Structure of porcine pancreatic phospholipase A2 at 2.6 A resolution and comparison with bovine phospholipase A2. Authors: Dijkstra, B.W. / Renetseder, R. / Kalk, K.H. / Hol, W.G. / Drenth, J. #1:  Journal: Acta Crystallogr.,Sect.B / Year: 1982 Journal: Acta Crystallogr.,Sect.B / Year: 1982Title: The Structure of Bovine Pancreatic Prophospholipase A2 at 3.0 Angstroms Resolution Authors: Dijkstra, B.W. / Vannes, G.J.H. / Kalk, K.H. / Brandenburg, N.P. / Hol, W.G.J. / Drenth, J. #2:  Journal: J.Mol.Biol. / Year: 1981 Journal: J.Mol.Biol. / Year: 1981Title: Structure of Bovine Pancreatic Phospholipase A2 at 1.7 Angstroms Resolution Authors: Dijkstra, B.W. / Kalk, K.H. / Hol, W.G.J. / Drenth, J. #3:  Journal: Nature / Year: 1981 Journal: Nature / Year: 1981Title: Active Site and Catalytic Mechanism of Phospholipase A2 Authors: Dijkstra, B.W. / Drenth, J. / Kalk, K.H. #4:  Journal: J.Mol.Biol. / Year: 1978 Journal: J.Mol.Biol. / Year: 1978Title: Three-Dimensional Structure and Disulfide Bond Connections in Bovine Pancreatic Phospholipase A2 Authors: Dijkstra, B.W. / Drenth, J. / Kalk, K.H. / Vandermaelen, P.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
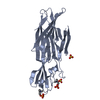
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1p2p.cif.gz 1p2p.cif.gz | 37 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1p2p.ent.gz pdb1p2p.ent.gz | 25.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1p2p.json.gz 1p2p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p2/1p2p https://data.pdbj.org/pub/pdb/validation_reports/p2/1p2p ftp://data.pdbj.org/pub/pdb/validation_reports/p2/1p2p ftp://data.pdbj.org/pub/pdb/validation_reports/p2/1p2p | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: SEE REMARK 8. / 2: SEE REMARK 4. | ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 14009.714 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | Compound details | THE TYPE CLASSIFICATIONS GIVEN IN THE REMARK FIELD OF THE TURN RECORDS BELOW FOLLOWS THE ...THE TYPE CLASSIFICA | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.4 Å3/Da / Density % sol: 63.79 % | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 7.2 / Method: unknown | ||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 2.6 Å / Num. all: 6144 / Num. obs: 4603 / % possible obs: 74.9 % / Rmerge(I) obs: 0.058 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor Rwork: 0.241 / Highest resolution: 2.6 Å | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2.6 Å
| ||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 7 Å / Num. reflection obs: 4295 / Rfactor obs: 0.241 | ||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 21.5 Å2 | ||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




