+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ow0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human FcaRI bound to IgA1-Fc | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | IMMUNE SYSTEM / IgA1 / FcaRI / CD89 / antibody / Fc receptor / immunoglobulin-like domain | |||||||||
| Function / homology |  Function and homology information Function and homology informationIgA receptor activity / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / cellular response to interferon-alpha / Fc receptor signaling pathway / IgA binding / immune response-regulating signaling pathway / positive regulation of neutrophil apoptotic process / IgA immunoglobulin complex ...IgA receptor activity / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / cellular response to interferon-alpha / Fc receptor signaling pathway / IgA binding / immune response-regulating signaling pathway / positive regulation of neutrophil apoptotic process / IgA immunoglobulin complex / glomerular filtration / neutrophil activation / cellular response to granulocyte macrophage colony-stimulating factor stimulus / neutrophil mediated immunity / cellular response to interleukin-6 / IgG immunoglobulin complex / immunoglobulin complex, circulating / positive regulation of respiratory burst / tertiary granule membrane / ficolin-1-rich granule membrane / complement activation, classical pathway / Scavenging of heme from plasma / antigen binding / specific granule membrane / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / cellular response to type II interferon / cellular response to tumor necrosis factor / antibacterial humoral response / cellular response to lipopolysaccharide / blood microparticle / adaptive immune response / immune response / Neutrophil degranulation / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | |||||||||
 Authors Authors | Herr, A.B. / Ballister, E.R. / Bjorkman, P.J. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2003 Journal: Nature / Year: 2003Title: Insights into IgA-mediated immune responses from the crystal structures of human Fc-alpha-RI and its complex with IgA1-Fc Authors: Herr, A.B. / Ballister, E.R. / Bjorkman, P.J. #1:  Journal: J.Mol.Biol. / Year: 2003 Journal: J.Mol.Biol. / Year: 2003Title: Bivalent Binding of IgA1 to FcaRI Suggests a Mechanism for Cytokine Activation of IgA Phagocytosis Authors: Herr, A.B. / White, C.L. / Milburn, C. / Wu, C. / Bjorkman, P.J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ow0.cif.gz 1ow0.cif.gz | 165.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ow0.ent.gz pdb1ow0.ent.gz | 130.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ow0.json.gz 1ow0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ow/1ow0 https://data.pdbj.org/pub/pdb/validation_reports/ow/1ow0 ftp://data.pdbj.org/pub/pdb/validation_reports/ow/1ow0 ftp://data.pdbj.org/pub/pdb/validation_reports/ow/1ow0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 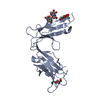 1ovzSC  1dn2S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological assembly state is a dimer of chains A and B, which are related by 2-fold NCS. / The biological assembly state is a monomer. |
- Components
Components
-Antibody / Protein , 2 types, 4 molecules ABCD
| #1: Antibody | Mass: 23226.391 Da / Num. of mol.: 2 / Fragment: Fc region Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: IGHA1 / Plasmid: pBJ5GS-MCS3 / Production host: Homo sapiens (human) / Gene: IGHA1 / Plasmid: pBJ5GS-MCS3 / Production host:  #2: Protein | Mass: 23615.742 Da / Num. of mol.: 2 / Fragment: ectodomain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FCAR OR CD89 / Plasmid: pAcGP67 / Production host: Homo sapiens (human) / Gene: FCAR OR CD89 / Plasmid: pAcGP67 / Production host:  Trichoplusia ni (cabbage looper) / Strain (production host): High 5 insect cells / References: UniProt: P24071 Trichoplusia ni (cabbage looper) / Strain (production host): High 5 insect cells / References: UniProt: P24071 |
|---|
-Sugars , 6 types, 10 molecules 
| #3: Polysaccharide | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D- ...N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source | ||||||
|---|---|---|---|---|---|---|---|
| #4: Polysaccharide | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D- ...N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source | ||||||
| #5: Polysaccharide | Source method: isolated from a genetically manipulated source #6: Polysaccharide | Source method: isolated from a genetically manipulated source #7: Polysaccharide | Source method: isolated from a genetically manipulated source #8: Sugar | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.93 Å3/Da / Density % sol: 69.3 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 296 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: magnesium sulfate heptahydrate, MES, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 296K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-1 / Wavelength: 0.95 Å / Beamline: BL9-1 / Wavelength: 0.95 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: May 4, 2002 |
| Radiation | Monochromator: single crystal Si(311) bent monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.95 Å / Relative weight: 1 |
| Reflection | Resolution: 3.1→30 Å / Num. all: 27807 / Num. obs: 27262 / % possible obs: 95.8 % / Observed criterion σ(I): -3 / Redundancy: 7.7 % / Biso Wilson estimate: 73.2 Å2 / Rmerge(I) obs: 0.089 / Net I/σ(I): 12 |
| Reflection shell | Resolution: 3.1→3.21 Å / Redundancy: 1.9 % / Rmerge(I) obs: 0.31 / Mean I/σ(I) obs: 2.4 / Num. unique all: 1998 / % possible all: 71.6 |
| Reflection | *PLUS Num. obs: 27807 / Num. measured all: 213861 |
| Reflection shell | *PLUS % possible obs: 71.6 % / Rmerge(I) obs: 0.31 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRIES 1DN2, 1OVZ Resolution: 3.1→29.69 Å / Rfactor Rfree error: 0.008 / Isotropic thermal model: GROUP / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 53.6167 Å2 / ksol: 0.292762 e/Å3 | |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 98.2 Å2
| |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.1→29.69 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: CONSTR | |||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.1→3.21 Å / Rfactor Rfree error: 0.041 / Total num. of bins used: 10
| |||||||||||||||||||||||||
| Xplor file |
| |||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 3.1 Å / Lowest resolution: 30 Å / Rfactor Rwork: 0.251 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj
















