[English] 日本語
 Yorodumi
Yorodumi- PDB-1oj5: Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1oj5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTIONAL COACTIVATOR / COMPLEX / LXXLL MOTIF / TRANSCRIPTIONAL REGULATION / STAT6 / PAS DOMAIN / IL-4 STAT | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mast cell proliferation / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / SUMOylation of transcription cofactors / mammary gland morphogenesis / Recycling of bile acids and salts / Synthesis of bile acids and bile salts / positive regulation of isotype switching to IgE isotypes / negative regulation of type 2 immune response / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol ...regulation of mast cell proliferation / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / SUMOylation of transcription cofactors / mammary gland morphogenesis / Recycling of bile acids and salts / Synthesis of bile acids and bile salts / positive regulation of isotype switching to IgE isotypes / negative regulation of type 2 immune response / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Synthesis of bile acids and bile salts via 27-hydroxycholesterol / Endogenous sterols / HATs acetylate histones / T-helper 1 cell lineage commitment / Regulation of lipid metabolism by PPARalpha / STAT6-mediated induction of chemokines / Cytoprotection by HMOX1 / interleukin-4-mediated signaling pathway / Estrogen-dependent gene expression / isotype switching to IgE isotypes / labyrinthine layer morphogenesis / positive regulation of transcription from RNA polymerase II promoter by galactose / regulation of thyroid hormone receptor signaling pathway / positive regulation of female receptivity / mammary gland epithelial cell proliferation / male mating behavior / hypothalamus development / histone acetyltransferase activity / cellular response to Thyroglobulin triiodothyronine / progesterone receptor signaling pathway / growth hormone receptor signaling pathway via JAK-STAT / cell surface receptor signaling pathway via JAK-STAT / response to retinoic acid / estrous cycle / nuclear retinoid X receptor binding / histone acetyltransferase / estrogen receptor signaling pathway / lactation / positive regulation of adipose tissue development / peroxisome proliferator activated receptor signaling pathway / positive regulation of neuron differentiation / regulation of cellular response to insulin stimulus / Downstream signal transduction / response to progesterone / cerebellum development / nuclear estrogen receptor binding / RNA polymerase II transcription regulatory region sequence-specific DNA binding / hippocampus development / defense response / cerebral cortex development / mRNA transcription by RNA polymerase II / response to peptide hormone / RNA polymerase II transcription regulator complex / male gonad development / transcription coactivator binding / cytokine-mediated signaling pathway / response to estradiol / regulation of cell population proliferation / positive regulation of cold-induced thermogenesis / DNA-binding transcription activator activity, RNA polymerase II-specific / Interleukin-4 and Interleukin-13 signaling / protein phosphatase binding / transcription regulator complex / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription coactivator activity / protein dimerization activity / positive regulation of apoptotic process / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / chromatin binding / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / chromatin / protein-containing complex binding / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / OTHER / Resolution: 2.21 Å X-RAY DIFFRACTION / OTHER / Resolution: 2.21 Å | ||||||
 Authors Authors | Razeto, A. / Ramakrishnan, V. / Giller, K. / Lakomek, N. / Carlomagno, T. / Griesinger, C. / Lodrini, M. / Litterst, C.M. / Pftizner, E. / Becker, S. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2004 Journal: J.Mol.Biol. / Year: 2004Title: Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain Authors: Razeto, A. / Ramakrishnan, V. / Litterst, C.M. / Giller, K. / Griesinger, C. / Carlomagno, T. / Lakomek, N. / Heimburg, T. / Lodrini, M. / Pfitzner, E. / Becker, S. #1: Journal: J.Biol.Chem. / Year: 2002 Title: An Lxxll Motif in the Transactivation Domain of Stat6 Mediates Recruitment of Ncoa-1/Src-1 Authors: Litterst, C.M. / Pfitzner, E. #2: Journal: J.Biol.Chem. / Year: 2001 Title: Transcriptional Activation by Stat6 Requires the Direct Interaction with Ncoa-1 Authors: Litterst, C.M. / Pfitzner, E. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. | ||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR PROVIDED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1oj5.cif.gz 1oj5.cif.gz | 40.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1oj5.ent.gz pdb1oj5.ent.gz | 27.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1oj5.json.gz 1oj5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oj/1oj5 https://data.pdbj.org/pub/pdb/validation_reports/oj/1oj5 ftp://data.pdbj.org/pub/pdb/validation_reports/oj/1oj5 ftp://data.pdbj.org/pub/pdb/validation_reports/oj/1oj5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 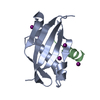
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | THIS BIOMOLECULE IS A HETERODIMERIC COMPLEX OF A PROTEINCHAIN WITH A PEPTIDE |
- Components
Components
| #1: Protein | Mass: 14811.774 Da / Num. of mol.: 1 / Fragment: NCO-A1 PAS-B DOMAIN, RESIDUES 257-385 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   | ||||||
|---|---|---|---|---|---|---|---|
| #2: Protein/peptide | Mass: 1610.865 Da / Num. of mol.: 1 / Fragment: STAT6 LXXLL MOTIF, RESIDUES 795-808 / Source method: obtained synthetically / Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: P42226 HOMO SAPIENS (human) / References: UniProt: P42226 | ||||||
| #3: Chemical | ChemComp-IOD / #4: Water | ChemComp-HOH / | Compound details | ENGINEERED | Sequence details | RESIDUES A254-A256 ARE FROM THE VECTOR PET16BTEV | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7 / Details: 20 % PEG3350, 0.2 M LICL, pH 7.00 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / pH: 7 / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: SIEMENS M18X / Wavelength: 1.5418 ROTATING ANODE / Type: SIEMENS M18X / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Mar 24, 2003 / Details: OSMIC MIRRORS CMF12-38CU6 |
| Radiation | Monochromator: OSMIC MIRRORS CMF12-38CU6 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.21→19.21 Å / Num. obs: 8138 / % possible obs: 99.7 % / Redundancy: 16.8 % / Biso Wilson estimate: 28.7 Å2 / Rmerge(I) obs: 0.089 / Net I/σ(I): 29.9 |
| Reflection shell | Resolution: 2.21→2.25 Å / Redundancy: 16 % / Rmerge(I) obs: 0.3254 / Mean I/σ(I) obs: 8.71 / % possible all: 98.1 |
| Reflection | *PLUS Highest resolution: 2.21 Å / Lowest resolution: 19.2 Å / Redundancy: 16.8 % / Num. measured all: 137915 / Rmerge(I) obs: 0.089 |
| Reflection shell | *PLUS % possible obs: 98.1 % / Redundancy: 16 % / Rmerge(I) obs: 0.325 / Mean I/σ(I) obs: 8.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: OTHER / Resolution: 2.21→19.21 Å / Cor.coef. Fo:Fc: 0.953 / Cor.coef. Fo:Fc free: 0.928 / SU B: 2.825 / SU ML: 0.075 / Cross valid method: THROUGHOUT / ESU R: 0.195 / ESU R Free: 0.178 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE LAST RUN OF REFINEMENT WAS PERFORMED WITH ALL THE REFLECTIONS. THE R FREE REPORTED ABOVE IS REFERRED TO THE PREVIOUS RUN OF REFINEMENT. ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE LAST RUN OF REFINEMENT WAS PERFORMED WITH ALL THE REFLECTIONS. THE R FREE REPORTED ABOVE IS REFERRED TO THE PREVIOUS RUN OF REFINEMENT. SIDE CHAINS WITH POOR ELECTRON DENSITY WERE ASSIGNED OCCUPANCY 0.5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL PLUS MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 32.92 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.21→19.21 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj




















