+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ocp | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF OCT3 POU-HOMEODOMAIN | ||||||
 Components Components | OCT-3 | ||||||
 Keywords Keywords | DNA BINDING PROTEIN / DNA-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationectodermal cell fate commitment / response to benzoic acid / mesodermal cell fate commitment / HMG box domain binding / trophectodermal cell fate commitment / regulation of asymmetric cell division / endodermal cell fate specification / blastocyst growth / POU domain binding / endodermal cell fate commitment ...ectodermal cell fate commitment / response to benzoic acid / mesodermal cell fate commitment / HMG box domain binding / trophectodermal cell fate commitment / regulation of asymmetric cell division / endodermal cell fate specification / blastocyst growth / POU domain binding / endodermal cell fate commitment / germ-line stem cell population maintenance / trophectodermal cell differentiation / negative regulation of calcium ion-dependent exocytosis / female germ cell nucleus / germ cell nucleus / RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding / stem cell population maintenance / cytokine binding / somatic stem cell population maintenance / negative regulation of cell differentiation / blastocyst development / cell fate commitment / positive regulation of transcription initiation by RNA polymerase II / response to retinoic acid / cis-regulatory region sequence-specific DNA binding / : / transcription repressor complex / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / telomere maintenance / cellular response to leukemia inhibitory factor / male germ cell nucleus / stem cell differentiation / chromatin DNA binding / nuclear receptor activity / DNA-binding transcription activator activity, RNA polymerase II-specific / transcription regulator complex / gene expression / sequence-specific DNA binding / DNA-binding transcription factor binding / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / transcription cis-regulatory region binding / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of gene expression / negative regulation of DNA-templated transcription / ubiquitin protein ligase binding / chromatin binding / positive regulation of gene expression / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / positive regulation of DNA-templated transcription / nucleolus / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Morita, E.H. / Hayashi, F. / Shirakawa, M. / Kyogoku, Y. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1995 Journal: Protein Sci. / Year: 1995Title: Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy Authors: Morita, E.H. / Shirakawa, M. / Hayashi, F. / Imagawa, M. / Kyogoku, Y. #1:  Journal: FEBS Lett. / Year: 1993 Journal: FEBS Lett. / Year: 1993Title: Secondary Structure of the Oct-3 POU Homeodomain as Determined by 1H-15N NMR Spectroscopy Authors: Morita, E.H. / Shirakawa, M. / Hayashi, F. / Imagawa, K. / Kyogoku, Y. | ||||||
| History |
|
- Structure visualization
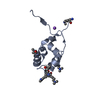
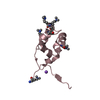
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ocp.cif.gz 1ocp.cif.gz | 452.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ocp.ent.gz pdb1ocp.ent.gz | 378.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ocp.json.gz 1ocp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ocp_validation.pdf.gz 1ocp_validation.pdf.gz | 344.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ocp_full_validation.pdf.gz 1ocp_full_validation.pdf.gz | 490.3 KB | Display | |
| Data in XML |  1ocp_validation.xml.gz 1ocp_validation.xml.gz | 27.9 KB | Display | |
| Data in CIF |  1ocp_validation.cif.gz 1ocp_validation.cif.gz | 44.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oc/1ocp https://data.pdbj.org/pub/pdb/validation_reports/oc/1ocp ftp://data.pdbj.org/pub/pdb/validation_reports/oc/1ocp ftp://data.pdbj.org/pub/pdb/validation_reports/oc/1ocp | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 8029.471 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene (production host): T7 GENE 1 UNDER CONTROL OF THE LAC UV5 PROMOTER Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement | ||||||||||||
| NMR ensemble | Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj


