[English] 日本語
 Yorodumi
Yorodumi- PDB-1o15: THEOPHYLLINE-BINDING RNA IN COMPLEX WITH THEOPHYLLINE, NMR, REGUL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1o15 | ||||||
|---|---|---|---|---|---|---|---|
| Title | THEOPHYLLINE-BINDING RNA IN COMPLEX WITH THEOPHYLLINE, NMR, REGULARIZED MEAN STRUCTURE, REFINEMENT WITH TORSION ANGLE AND BASE-BASE POSITIONAL DATABASE POTENTIALS AND DIPOLAR COUPLINGS | ||||||
 Components Components | THEOPHYLLINE-BINDING RNA | ||||||
 Keywords Keywords | RNA / RIBONUCLEIC ACID | ||||||
| Function / homology | THEOPHYLLINE / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Clore, G.M. / Kuszewski, J. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2003 Journal: J.Am.Chem.Soc. / Year: 2003Title: Improving the Accuracy of NMR Structures of RNA by Means of Conformational Database Potentials of Mean Force as Assessed by Complete Dipolar Coupling Cross-Validation Authors: Clore, G.M. / Kuszewski, J. #1:  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: Interlocking Structural Motifs Mediate Molecular Discrimination by a Theophylline-Binding RNA Authors: Zimmermann, G.R. / Jenison, R.D. / Wick, C.L. / Simorre, J.P. / Pardi, A. #2:  Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Refinement of Local and Long Range Structural Order in Theophylline-Binding RNA Using Using 13C-1H Residual Dipolar Couplings and Restrained Molecular Dynamics. Authors: Sibille, N. / Pardi, A. / Simorre, J.P. / Blackledge, M. #3:  Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Improving the Accuracy of NMR Structures of DNA by Means of a Database Potential of Mean Force Describing Base-Base Positional Interactions. Authors: Kuszewski, J. / Schwieters, C. / Clore, G.M. | ||||||
| History |
| ||||||
| Remark 8 | THE RESTRAINTS USED FOR DETERMINATION OF THIS STRUCTURE ARE SUMMARIZED AS FOLLOWS: DISTANCE ... THE RESTRAINTS USED FOR DETERMINATION OF THIS STRUCTURE ARE SUMMARIZED AS FOLLOWS: DISTANCE RESTRAINTS 30 INTRARESIDUE NOES 86 SEQUENTIAL NOES 17 MEDIUM RANGE (1 < |i-j|<5) NOES 90 LONG RANGE (|i-j|>=5) NOES 52 DISTANCES FOR 8 W-C BASE PAIRS and 1 G-U WOBBLE PAIR TOTAL: 223 LOOSE TORSION ANGLE RESTRAINTS 31 DELTA 33 CHI 9 ALPHA, 9 BETA, 10 GAMMA, 9 EPSILON AND 9 ZETA TOTAL: 110. 13C-1H DIPOLAR COUPLINGS 55 SUGAR 46 BASE TOTAL 101 |
- Structure visualization
Structure visualization
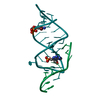
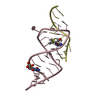
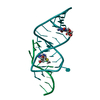
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1o15.cif.gz 1o15.cif.gz | 33 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1o15.ent.gz pdb1o15.ent.gz | 23.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1o15.json.gz 1o15.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o1/1o15 https://data.pdbj.org/pub/pdb/validation_reports/o1/1o15 ftp://data.pdbj.org/pub/pdb/validation_reports/o1/1o15 ftp://data.pdbj.org/pub/pdb/validation_reports/o1/1o15 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 10638.409 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: CONSENSUS SEQUENCE OF THE THEOPHYLLINE BINDING RNA APTAMER FLANKED BY A FOUR BASE-PAIR STEM, A THREE BASE-PAIR STEM, AND CAPPED BY A GAAA TETRALOOP |
|---|---|
| #2: Chemical | ChemComp-TEP / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: THE DIPOLAR COUPLINGS WERE DIVIDED INTO 10 PAIRS OF WORKING AND TEST DATA SETS CHOSEN AT RANDOM AND PARTITIONED IN A RATIO OF 70% (WORKING) AND 30% (TEST). 25 SIMULATED ANNEALING STRUCTURES ...Text: THE DIPOLAR COUPLINGS WERE DIVIDED INTO 10 PAIRS OF WORKING AND TEST DATA SETS CHOSEN AT RANDOM AND PARTITIONED IN A RATIO OF 70% (WORKING) AND 30% (TEST). 25 SIMULATED ANNEALING STRUCTURES WERE CALCULATED FOR EACH PAIR, RESULTING IN A TOTAL OF 250 STRUCTURES. THE STRUCTURE REPORTED HERE IS OBTAINED BY RESTRAINED REGULARIZED OF THE MEAN COORDINATES OF THE ENSEMBLE OF 250 INDIVIDUAL SIMULATED ANNEALING STRUCTURES CALCULATED WITH COMPLETE DIPOLAR COUPLING CROSS-VALIDATION. FOR THE ENSEMBLE OF 250 STRUCTURES THE WORKING DIPOLAR COUPLING R-FACTOR IS 8.6+/-0.5% AND THE FREE DIPOLAR COUPLING R-FACTOR IS 26.8+/-2.8%. |
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| NMR software | Name:  X-PLOR NIH / Version: (HTTP://NMR.CIT.NIH.GOV/XPLOR_NIH) / Developer: CLORE, KUSZEWSKI, SCHWIETERS, TJANDRA / Classification: refinement X-PLOR NIH / Version: (HTTP://NMR.CIT.NIH.GOV/XPLOR_NIH) / Developer: CLORE, KUSZEWSKI, SCHWIETERS, TJANDRA / Classification: refinement |
|---|---|
| Refinement | Software ordinal: 1 Details: THE STRUCTURE IS BASED ON A TOTAL OF 223 NOE, 52 DISTANCES FOR WATSON-CRICK HYDROGEN BONDS, AND 110 TORSION ANGLE RESTRAINTS, 101 CH DIPOLAR COUPLING RESTRAINTS. THE EXPERIMENTAL RESTRAINTS ...Details: THE STRUCTURE IS BASED ON A TOTAL OF 223 NOE, 52 DISTANCES FOR WATSON-CRICK HYDROGEN BONDS, AND 110 TORSION ANGLE RESTRAINTS, 101 CH DIPOLAR COUPLING RESTRAINTS. THE EXPERIMENTAL RESTRAINTS ARE ESSENTIALLY THE SAME AS THOSE LISTED IN N.SIBILLE,A.PARDI,J.P.SIMORRE,M.BLACKLEDGE J.AM.CHEM.SOC. V 123, 12135 2001. THE NON-BONDED CONTACTS ARE REPRESENTED BY A QUARTIC VAN DER WAALS REPULSION TERM, A BASE-BASE POSITIONING DATABASE POTENTIAL OF MEAN FORCE, AND A TORSION ANGLE DATABASE POTENTIAL OF MEAN FORCE. |
| NMR ensemble | Conformer selection criteria: RESTRAINED REGULARIZED MEAN STRUCTURE Conformers calculated total number: 250 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller









 PDBj
PDBj