[English] 日本語
 Yorodumi
Yorodumi- PDB-1lmv: Solution structure of the unmodified U2 snRNA-intron branch site ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lmv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of the unmodified U2 snRNA-intron branch site helix from S. cerevisiae | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA / U2 snRNA / branch site / solution structure / A-form helix | ||||||
| Function / homology | RNA Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / torsion angle molecular dynamics, congugate gradient minimization | ||||||
 Authors Authors | Newby, M.I. / Greenbaum, N.L. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2002 Journal: Nat.Struct.Biol. / Year: 2002Title: Sculpting of the Spliceosomal Branch Site Recognition Motif by a Conserved Pseudouridine Authors: Newby, M.I. / Greenbaum, N.L. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE Position 35 in U2 snRNA sequences from S. cerevisiae is a pseudouridine. This corresponds ...SEQUENCE Position 35 in U2 snRNA sequences from S. cerevisiae is a pseudouridine. This corresponds to position 5 on strand A, which is a uridine in 1LMV, and a pseudouridine in 1LPW. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lmv.cif.gz 1lmv.cif.gz | 123.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lmv.ent.gz pdb1lmv.ent.gz | 102.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lmv.json.gz 1lmv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lm/1lmv https://data.pdbj.org/pub/pdb/validation_reports/lm/1lmv ftp://data.pdbj.org/pub/pdb/validation_reports/lm/1lmv ftp://data.pdbj.org/pub/pdb/validation_reports/lm/1lmv | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 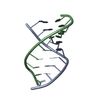
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 2912.775 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: unmodified U2 snRNA. Region of U2 snRNA that pairs with the intron branch site during spliceosome assembly (nucleotides 34-39 in yeast U2 snRNA). Source: (natural)  |
|---|---|
| #2: RNA chain | Mass: 3104.925 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: Intron branch site sequence. Region of the pre-mRNA intron that contains the conserved branch site sequence, and pairs with U2 snRNA during spliceosome assembly. Source: (natural)  |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. NOESY spectra were collected at many different mixing times to assess the buildup rate of each NOE, thereby resolving ...Text: This structure was determined using standard 2D homonuclear techniques. NOESY spectra were collected at many different mixing times to assess the buildup rate of each NOE, thereby resolving some of the spectral overlap. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 720 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle molecular dynamics, congugate gradient minimization Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 300 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller







 PDBj
PDBj X-PLOR
X-PLOR