[English] 日本語
 Yorodumi
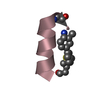
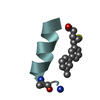
Yorodumi- PDB-1kmr: Solution NMR Structure of Surfactant Protein B (11-25) (SP-B11-25) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kmr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR Structure of Surfactant Protein B (11-25) (SP-B11-25) | ||||||
 Components Components | PULMONARY SURFACTANT-ASSOCIATED PROTEIN B | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN / HELIX | ||||||
| Function / homology |  Function and homology information Function and homology informationDefective pro-SFTPB causes SMDP1 and RDS / multivesicular body lumen / lamellar body / alveolar lamellar body / Defective CSF2RB causes SMDP5 / Defective CSF2RA causes SMDP4 / sphingolipid metabolic process / clathrin-coated endocytic vesicle / respiratory gaseous exchange by respiratory system / Surfactant metabolism ...Defective pro-SFTPB causes SMDP1 and RDS / multivesicular body lumen / lamellar body / alveolar lamellar body / Defective CSF2RB causes SMDP5 / Defective CSF2RA causes SMDP4 / sphingolipid metabolic process / clathrin-coated endocytic vesicle / respiratory gaseous exchange by respiratory system / Surfactant metabolism / multivesicular body / animal organ morphogenesis / lysosome / endoplasmic reticulum membrane / extracellular space / extracellular region Similarity search - Function | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Kurutz, J.W. / Lee, K.Y.C. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: NMR structure of lung surfactant peptide SP-B(11-25). Authors: Kurutz, J.W. / Lee, K.Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kmr.cif.gz 1kmr.cif.gz | 79.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kmr.ent.gz pdb1kmr.ent.gz | 54.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kmr.json.gz 1kmr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/km/1kmr https://data.pdbj.org/pub/pdb/validation_reports/km/1kmr ftp://data.pdbj.org/pub/pdb/validation_reports/km/1kmr ftp://data.pdbj.org/pub/pdb/validation_reports/km/1kmr | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1702.183 Da / Num. of mol.: 1 Fragment: SEQUENCE DATABASE RESIDUES 211-225, NUMBERED 11-25 Source method: obtained synthetically Details: THE PROTEIN WAS CHEMICALLY SYNTHESIZED. THE SEQUENCE OF THE PROTEIN IS NATURALLY FOUND IN HOMO SAPIENS (HUMANS). References: UniProt: P07988 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: 2D NOESY |
| NMR details | Text: WET water suppression |
- Sample preparation
Sample preparation
| Details | Contents: 1.25 mM peptide, 0.1 mM DSS-d6 Solvent system: Partially deuterated methanol (CD3OH), 98% D |
|---|---|
| Sample conditions | Ionic strength: no additional salt or buffer / pH: 5.5 / Pressure: ambient / Temperature: 278 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 78 / Conformers submitted total number: 17 |
 Movie
Movie Controller
Controller








 PDBj
PDBj