Entry Database : PDB / ID : 1kbfTitle Solution Structure of the Cysteine-Rich C1 Domain of Kinase Suppressor of Ras Kinase suppressor of Ras 1 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Method / Authors Zhou, M. / Horita, D.A. / Waugh, D.S. / Byrd, R.A. / Morrison, D.K. Journal : J.Mol.Biol. / Year : 2002Title : Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).Authors : Zhou, M. / Horita, D.A. / Waugh, D.S. / Byrd, R.A. / Morrison, D.K. History Deposition Nov 6, 2001 Deposition site / Processing site Revision 1.0 Jan 23, 2002 Provider / Type Revision 1.1 Apr 27, 2008 Group Revision 1.2 Jul 13, 2011 Group Revision 1.3 Dec 25, 2019 Group / Derived calculations / OtherCategory database_2 / pdbx_database_related ... database_2 / pdbx_database_related / pdbx_database_status / pdbx_struct_assembly / pdbx_struct_oper_list / struct_ref_seq_dif Item / _struct_ref_seq_dif.detailsRevision 2.0 Feb 26, 2020 Group Atomic model / Author supporting evidence ... Atomic model / Author supporting evidence / Data collection / Database references / Derived calculations / Experimental preparation / Other / Refinement description / Source and taxonomy / Structure summary Category atom_site / diffrn ... atom_site / diffrn / diffrn_radiation / diffrn_radiation_wavelength / entity / entity_name_com / entity_src_gen / pdbx_database_status / pdbx_entity_instance_feature / pdbx_entry_details / pdbx_nmr_exptl / pdbx_nmr_exptl_sample / pdbx_nmr_exptl_sample_conditions / pdbx_nmr_refine / pdbx_nmr_sample_details / pdbx_struct_conn_angle / pdbx_struct_sheet_hbond / pdbx_validate_torsion / pdbx_version / struct / struct_conn / struct_keywords / struct_ref_seq_dif / struct_sheet / struct_sheet_order / struct_sheet_range / struct_site Item _atom_site.Cartn_x / _atom_site.Cartn_y ... _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _entity.pdbx_description / _entity.pdbx_ec / _entity.pdbx_fragment / _entity_src_gen.gene_src_common_name / _entity_src_gen.pdbx_beg_seq_num / _entity_src_gen.pdbx_end_seq_num / _entity_src_gen.pdbx_gene_src_gene / _entity_src_gen.pdbx_seq_type / _pdbx_database_status.SG_entry / _pdbx_database_status.status_code_mr / _pdbx_nmr_exptl_sample_conditions.ionic_strength / _pdbx_nmr_exptl_sample_conditions.ionic_strength_units / _pdbx_nmr_exptl_sample_conditions.label / _pdbx_nmr_exptl_sample_conditions.pH_units / _pdbx_nmr_sample_details.contents / _pdbx_nmr_sample_details.label / _pdbx_nmr_sample_details.type / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _pdbx_struct_sheet_hbond.sheet_id / _struct.pdbx_CASP_flag / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_keywords.text / _struct_ref_seq_dif.details / _struct_sheet.id / _struct_sheet_order.sheet_id / _struct_sheet_range.sheet_id / _struct_site.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id Description / Provider / Type Revision 2.1 Apr 17, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Other Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_mr / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id Revision 2.2 Oct 30, 2024 Group / Structure summaryCategory / pdbx_entry_details / pdbx_modification_featureItem / _pdbx_entry_details.has_protein_modification
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 Authors
Authors Citation
Citation Journal: J.Mol.Biol. / Year: 2002
Journal: J.Mol.Biol. / Year: 2002 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1kbf.cif.gz
1kbf.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1kbf.ent.gz
pdb1kbf.ent.gz PDB format
PDB format 1kbf.json.gz
1kbf.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/kb/1kbf
https://data.pdbj.org/pub/pdb/validation_reports/kb/1kbf ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kbf
ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kbf
 Links
Links Assembly
Assembly
 Components
Components

 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller






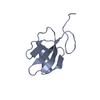


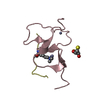


 PDBj
PDBj




 HSQC
HSQC