[English] 日本語
 Yorodumi
Yorodumi- PDB-1k76: Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1k76 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | ||||||
 Components Components | SEX MUSCLE ABNORMAL PROTEIN 5 | ||||||
 Keywords Keywords | SIGNALING PROTEIN / all beta protein | ||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by SCF-KIT / Regulation of KIT signaling / GRB2 events in ERBB2 signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / NCAM signaling for neurite out-growth / : / PI-3K cascade:FGFR1 / FRS-mediated FGFR1 signaling / SHC-mediated cascade:FGFR2 / FRS-mediated FGFR2 signaling ...Signaling by SCF-KIT / Regulation of KIT signaling / GRB2 events in ERBB2 signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / NCAM signaling for neurite out-growth / : / PI-3K cascade:FGFR1 / FRS-mediated FGFR1 signaling / SHC-mediated cascade:FGFR2 / FRS-mediated FGFR2 signaling / SHC-mediated cascade:FGFR3 / FRS-mediated FGFR3 signaling / FRS-mediated FGFR4 signaling / : / PI-3K cascade:FGFR4 / Insulin receptor signalling cascade / MET activates RAS signaling / : / RHOU GTPase cycle / FLT3 Signaling / PIP3 activates AKT signaling / GRB2 events in EGFR signaling / SHC1 events in EGFR signaling / PI3K events in ERBB2 signaling / Regulation of actin dynamics for phagocytic cup formation / EGFR Transactivation by Gastrin / RHO GTPases Activate WASPs and WAVEs / RAF/MAP kinase cascade / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / Downstream signal transduction / : / : / regulation of nematode larval development / Negative regulation of MET activity / EGFR downregulation / Cargo recognition for clathrin-mediated endocytosis / Regulation of signaling by CBL / regulation of vulval development / Clathrin-mediated endocytosis / regulation of cell projection organization / male genitalia development / COP9 signalosome / epidermal growth factor receptor binding / muscle organ development / regulation of MAPK cascade / phosphotyrosine residue binding / regulation of cell migration / epidermal growth factor receptor signaling pathway / signal transduction / nucleoplasm / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / distance geometry simulated annealing, molecular dynamics | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Ferreon, J. / Volk, D. / Luxon, B. / Gorenstein, D. / Hilser, V. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2003 Journal: Biochemistry / Year: 2003Title: Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble of Sem-5 C-Terminal SH3 Domain Authors: Ferreon, J. / Volk, D. / Luxon, B. / Gorenstein, D. / Hilser, V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1k76.cif.gz 1k76.cif.gz | 32.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1k76.ent.gz pdb1k76.ent.gz | 21.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1k76.json.gz 1k76.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k7/1k76 https://data.pdbj.org/pub/pdb/validation_reports/k7/1k76 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/1k76 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/1k76 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
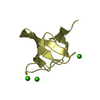
| Deposited unit | 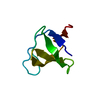
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7237.967 Da / Num. of mol.: 1 / Fragment: SH3 domain (Residues 155-214) / Mutation: C57A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||
| NMR details | Text: The structure was determined using triple-resonance NMR spectroscopy |
- Sample preparation
Sample preparation
| Details | Contents: 13C,15N 1mM Sem-5 SH3 domain 50 mM sodium acetate buffer, 10 mM CaCl2, 100 mM NaCl,pH 4.8, 90%h20, 10%d20 Solvent system: 90%h20, 10%d20 |
|---|---|
| Sample conditions | Ionic strength: 100 mM NaCl / pH: 4.8 / Pressure: ambient / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 400 MHz |
- Processing
Processing
| Software | Name: CNS / Classification: refinement | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||||||||||
| Refinement | Method: distance geometry simulated annealing, molecular dynamics Software ordinal: 1 | ||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations,structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller










 PDBj
PDBj HSQC
HSQC