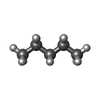[English] 日本語
 Yorodumi
Yorodumi- PDB-1in2: Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1in2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | ||||||
 Components Components | IGFBP-1 antagonist | ||||||
 Keywords Keywords | ANTAGONIST / covalently constrained helix | ||||||
| Function / homology | PENTANE Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / restrained molecular dynamics | ||||||
 Authors Authors | Skelton, N.J. / Chen, Y.M. / Dubree, N. / Quan, C. / Jackson, D.Y. / Cochran, A.G. / Zobel, K. / Deshayes, K. / Baca, M. / Pisabarro, M.T. / Lowman, H.B. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2001 Journal: Biochemistry / Year: 2001Title: Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1. Authors: Skelton, N.J. / Chen, Y.M. / Dubree, N. / Quan, C. / Jackson, D.Y. / Cochran, A. / Zobel, K. / Deshayes, K. / Baca, M. / Pisabarro, M.T. / Lowman, H.B. #1:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Molecular Mimics of Insulin-like Growth Factor 1 (IGF-1) for Inhibiting IGF-1: IGF-Binding Protein Interactions Authors: Lowman, H.B. / Chen, Y.M. / Skelton, N.J. / Mortensen, D.L. / Tomlinson, E.E. / Sadick, M.D. / Robinson, I.C. / Clark, R.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1in2.cif.gz 1in2.cif.gz | 92 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1in2.ent.gz pdb1in2.ent.gz | 63 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1in2.json.gz 1in2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1in2_validation.pdf.gz 1in2_validation.pdf.gz | 356.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1in2_full_validation.pdf.gz 1in2_full_validation.pdf.gz | 383.3 KB | Display | |
| Data in XML |  1in2_validation.xml.gz 1in2_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  1in2_validation.cif.gz 1in2_validation.cif.gz | 10.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/in/1in2 https://data.pdbj.org/pub/pdb/validation_reports/in/1in2 ftp://data.pdbj.org/pub/pdb/validation_reports/in/1in2 ftp://data.pdbj.org/pub/pdb/validation_reports/in/1in2 | HTTPS FTP |
-Related structure data
| Related structure data |  1gjeC  1gjfC  1gjgC  1imwC  1in3C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 1642.879 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: The peptide was chemically synthesized. It was designed from sequence selected from a phage display library. |
|---|---|
| #2: Chemical | ChemComp-LNK / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: restrained molecular dynamics / Software ordinal: 1 Details: The structure was detemined on the basis of 138 NOE distance restraints and 11 dihedral angle restraints. The resulting ensemble had no restraint violations greater than 0.07 Angstroms or 1. ...Details: The structure was detemined on the basis of 138 NOE distance restraints and 11 dihedral angle restraints. The resulting ensemble had no restraint violations greater than 0.07 Angstroms or 1.4 deg. The mean restraint violation energy was 0.04 +/- 0.03 kcal/mol. | ||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 80 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller


 PDBj
PDBj