[English] 日本語
 Yorodumi
Yorodumi- PDB-1i0m: 1.05 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-F... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1i0m | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1.05 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-FLUOROETHYL THYMINE IN PLACE OF T6, HIGH RB-SALT | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / A-form double helix / modified sugar | Function / homology | RUBIDIUM ION / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.05 Å SYNCHROTRON / Resolution: 1.05 Å  Authors AuthorsTereshko, V. / Wilds, C.J. / Minasov, G. / Prakash, T.P. / Maier, M.A. / Howard, A. / Wawrzak, Z. / Manoharan, M. / Egli, M. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2001 Journal: Nucleic Acids Res. / Year: 2001Title: Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments. Authors: Tereshko, V. / Wilds, C.J. / Minasov, G. / Prakash, T.P. / Maier, M.A. / Howard, A. / Wawrzak, Z. / Manoharan, M. / Egli, M. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1i0m.cif.gz 1i0m.cif.gz | 30.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1i0m.ent.gz pdb1i0m.ent.gz | 19.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1i0m.json.gz 1i0m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i0/1i0m https://data.pdbj.org/pub/pdb/validation_reports/i0/1i0m ftp://data.pdbj.org/pub/pdb/validation_reports/i0/1i0m ftp://data.pdbj.org/pub/pdb/validation_reports/i0/1i0m | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1i0fC  1i0gC  1i0jC  1i0kC  1i0nC  1i0oC  1i0pC  1i0qC  1i0tC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 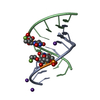
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3107.048 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.91 Å3/Da / Density % sol: 35.55 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6 Details: Rb salt, pH 6.0, VAPOR DIFFUSION, HANGING DROP at 298 K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 0.95 Å / Beamline: 17-ID / Wavelength: 0.95 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Nov 1, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.95 Å / Relative weight: 1 |
| Reflection | Resolution: 1.05→20 Å / Num. all: 22802 / Num. obs: 22802 / % possible obs: 100 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Rmerge(I) obs: 0.046 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.05→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.05→20 Å
| ||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | ||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller






 PDBj
PDBj





