[English] 日本語
 Yorodumi
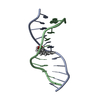
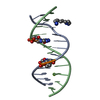
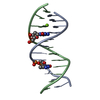
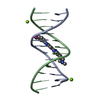
Yorodumi- PDB-1hx4: MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hx4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / 1R-trans-anti-BPh-N2-G adduct / carcinogen-dna adduct / benzo[c]phenanthrene dna adduct | ||||||
| Function / homology | Chem-BPJ / DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / torsion angle dynamics, simulated annealing, relaxation matrix refinements, energe minimization | ||||||
 Authors Authors | Patel, D.J. / Lin, C.H. / Geacintov, N.E. / Broyde, S. / Huang, X. / Kolbanovskii, A. / Hingerty, B.E. / Amin, S. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity. Authors: Lin, C.H. / Huang, X. / Kolbanovskii, A. / Hingerty, B.E. / Amin, S. / Broyde, S. / Geacintov, N.E. / Patel, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hx4.cif.gz 1hx4.cif.gz | 148.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hx4.ent.gz pdb1hx4.ent.gz | 117.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hx4.json.gz 1hx4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1hx4_validation.pdf.gz 1hx4_validation.pdf.gz | 411.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1hx4_full_validation.pdf.gz 1hx4_full_validation.pdf.gz | 480.3 KB | Display | |
| Data in XML |  1hx4_validation.xml.gz 1hx4_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  1hx4_validation.cif.gz 1hx4_validation.cif.gz | 15.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hx/1hx4 https://data.pdbj.org/pub/pdb/validation_reports/hx/1hx4 ftp://data.pdbj.org/pub/pdb/validation_reports/hx/1hx4 ftp://data.pdbj.org/pub/pdb/validation_reports/hx/1hx4 | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3254.138 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Synthesized from ABI-DNA Synthesizer |
|---|---|
| #2: DNA chain | Mass: 3454.258 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Synthesized from ABI-DNA Synthesizer |
| #3: Chemical | ChemComp-BPJ / ( |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: D2O NMR experiments were performed at 13 degree C, H2O NMR experiments were performed at 4 degree C |
- Sample preparation
Sample preparation
| Details | Contents: 1R-trans-anti-BPh (carcinogen) covalently bound to 11mer DNA duplex Solvent system: aqueous solution |
|---|---|
| Sample conditions | Ionic strength: 0.10 / pH: 6.5 / Pressure: 1 atm / Temperature: 277 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics, simulated annealing, relaxation matrix refinements, energe minimization Software ordinal: 1 Details: Nonexchangeable interproton distance restraints were obtained from the buildup of NOE cross peak volumes in NOESY data sets (55, 100, 150 and 200 ms mixing times) on the 1S-(-)-trans-anti- ...Details: Nonexchangeable interproton distance restraints were obtained from the buildup of NOE cross peak volumes in NOESY data sets (55, 100, 150 and 200 ms mixing times) on the 1S-(-)-trans-anti-(BPh)G*C and the 1R-(+)-trans-anti-(BPh)G*C 11-mer DNA duplex adducts in D2O buffer at 23 and 18 sC, respectively, and bounds were set between 15% and 25% of the calculated distances using the fixed cytidine H5-H6 reference distance of 2.45 . Non-stereospecific assignments were treated with r-6 averaging. Interproton distance restraints involving exchangeable protons of the same complex were obtained from NOESY spectra (90 and 150 ms mixing times) in H2O buffer at 4 sC with bounds set between 20% and 25% of the calculated distance using the thymine imino to adenine H2 reference distance of 2.91 across an A*T base pair. | ||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 9 / Conformers submitted total number: 9 |
 Movie
Movie Controller
Controller









 PDBj
PDBj







































 HSQC
HSQC