[English] 日本語
 Yorodumi
Yorodumi- PDB-1ho6: CHIMERIC ARABINONUCLEIC ACID (ANA) HAIRPIN WITH ANA/RNA HYBRID STEM -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ho6 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CHIMERIC ARABINONUCLEIC ACID (ANA) HAIRPIN WITH ANA/RNA HYBRID STEM | ||||||||||||||||||
 Components Components | DNA/RNA (5'-R(* Keywords KeywordsDNA / RNA / Arabinonucleic acid / Hairpin | Function / homology | DNA/RNA hybrid / DNA/RNA hybrid (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing |  Authors AuthorsDenisov, A.Y. / Gehring, K. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2001 Journal: Nucleic Acids Res. / Year: 2001Title: Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2'-fluoro-ANA/RNA and DNA/RNA hybrids. Authors: Denisov, A.Y. / Noronha, A.M. / Wilds, C.J. / Trempe, J.F. / Pon, R.T. / Gehring, K. / Damha, M.J. #1:  Journal: Nucleosides and Nucleotides / Year: 2001 Journal: Nucleosides and Nucleotides / Year: 2001Title: Properties of Arabinonucleic Acids (ANA & 2'F-ANA): Implications for the Design of Antisense Therapeutics that Invoke RNase H Cleavage of RNA Authors: Damha, M.J. / Noronha, A.M. / Wilds, C.J. / Trempe, J.F. / Denisov, A.Y. / Pon, R.T. / Gehring, K. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ho6.cif.gz 1ho6.cif.gz | 83.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ho6.ent.gz pdb1ho6.ent.gz | 51.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ho6.json.gz 1ho6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ho/1ho6 https://data.pdbj.org/pub/pdb/validation_reports/ho/1ho6 ftp://data.pdbj.org/pub/pdb/validation_reports/ho/1ho6 ftp://data.pdbj.org/pub/pdb/validation_reports/ho/1ho6 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 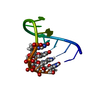
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA/RNA hybrid | Mass: 3768.350 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Solid phase synthesis, phosphoramidite chemistry |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||
| NMR details | Text: This structure was determined using standard 2D homo- and heteronuclear techniques: NOESY, DQF-COSY, TOCSY, H,C-HMQC and H,P-HetCOSY |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 7 / Pressure: 1 atm / Temperature: 288 K | |||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: Starting A- and B-type stem hairpin structures with randomly organized loops were refined by 14 ps restrained molecular dynamics (300-1000 K) with final enegry minimization | ||||||||||||
| NMR representative | Selection criteria: the hairpin stem structure closest to the average | ||||||||||||
| NMR ensemble | Conformer selection criteria: all calculated structures submitted Conformers calculated total number: 10 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller






 PDBj
PDBj


 X-PLOR
X-PLOR