[English] 日本語
 Yorodumi
Yorodumi- PDB-1hi7: NMR SOLUTION STRUCTURE OF THE DISULPHIDE-LINKED HOMODIMER OF HUMA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hi7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR SOLUTION STRUCTURE OF THE DISULPHIDE-LINKED HOMODIMER OF HUMAN TFF1, 10 STRUCTURES | ||||||
 Components Components | PS2 PROTEIN | ||||||
 Keywords Keywords | GROWTH FACTOR / CELL MOTILITY / TUMOR SUPPRESSOR / TREFOIL DOMAIN | ||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of gastrointestinal epithelium / response to iron ion / response to immobilization stress / growth factor activity / response to peptide hormone / Estrogen-dependent gene expression / cell population proliferation / carbohydrate metabolic process / cell differentiation / negative regulation of cell population proliferation ...maintenance of gastrointestinal epithelium / response to iron ion / response to immobilization stress / growth factor activity / response to peptide hormone / Estrogen-dependent gene expression / cell population proliferation / carbohydrate metabolic process / cell differentiation / negative regulation of cell population proliferation / extracellular space / extracellular region Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Williams, M.A. / Feeney, J. | ||||||
 Citation Citation |  Journal: FEBS Lett. / Year: 2001 Journal: FEBS Lett. / Year: 2001Title: The Solution Structure of the Disulphide-Linked Dimeric of the Human Trefoil Protein Tff1 Authors: Williams, M.A. / Westley, B.R. / May, F.E. / Feeney, J. #1:  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: High-Resolution Solution Structure of Human Pnr-2/ Ps2: A Single Trefoil Motif Protein Authors: Polshakov, V.I. / Williams, M.A. / Gargaro, A.R. / Frenkiel, T.A. / Westley, B.R. / Chadwick, M.P. / May, F.E.B. / Feeney, J. #2: Journal: Biochem.J. / Year: 1997 Title: Homodimerization and Hetero_Oligomerization of the Single Domain Trefoil Protein Pnr-2/Ps2 Through Cysteine 58 Authors: Chadwick, M.P. / May, F.E.B. / Westley, B.R. #3: Journal: Biochem.J. / Year: 1995 Title: Production and Comparison of Mature Single-Domain 'Trefoil' Peptides Pnr-2/Ps2 Cys58 and Pnr-2/Ps2 Ser58 Authors: Chadwick, M.P. / May, F.E.B. / Westley, B.R. #4: Journal: Eur.J.Biochem. / Year: 1995 Title: NMR-Based Structural Studies of the Pnr-2/Ps2 Single Domain Trefoil Peptide. Similarities to Porcine Spasmolytic Peptide and Evidence for a Monomeric Structure Authors: Polshakov, V.I. / Frenkiel, T.A. / Westley, B. / Chadwick, M. / May, F. / Carr, M.D. / Feeney, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
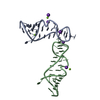
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hi7.cif.gz 1hi7.cif.gz | 348.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hi7.ent.gz pdb1hi7.ent.gz | 291 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hi7.json.gz 1hi7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1hi7_validation.pdf.gz 1hi7_validation.pdf.gz | 353.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1hi7_full_validation.pdf.gz 1hi7_full_validation.pdf.gz | 534 KB | Display | |
| Data in XML |  1hi7_validation.xml.gz 1hi7_validation.xml.gz | 49.6 KB | Display | |
| Data in CIF |  1hi7_validation.cif.gz 1hi7_validation.cif.gz | 64.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hi/1hi7 https://data.pdbj.org/pub/pdb/validation_reports/hi/1hi7 ftp://data.pdbj.org/pub/pdb/validation_reports/hi/1hi7 ftp://data.pdbj.org/pub/pdb/validation_reports/hi/1hi7 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6678.365 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: DISULPHIDE LINK BETWEEN C58 OF BOTH SUBUNITS / Source: (gene. exp.)  HOMO SAPIENS (human) / Tissue: EPITHELIAL / Cell: MCF-7, UACL / Cellular location: EXTRACELLULAR / Gene: TFF1 / Organ: BREAST, STOMACH / Cellular location (production host): CYTOPLASM / Gene (production host): TFF1 / Production host: HOMO SAPIENS (human) / Tissue: EPITHELIAL / Cell: MCF-7, UACL / Cellular location: EXTRACELLULAR / Gene: TFF1 / Organ: BREAST, STOMACH / Cellular location (production host): CYTOPLASM / Gene (production host): TFF1 / Production host:  Has protein modification | Y | Sequence details | 1PS2 SWS P04155 1 - 24 NOT IN ATOMS LIST | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TWO AND THREE DIMENSIONAL NMR EXPERIMENTS ON, RESPECTIVELY, UNLABELLED AND N-15 LABELLED TFF1 DIMER PROTEIN. MANY ASSIGN WERE MADE BY MAKING USE IF THE BETTER ...Text: THE STRUCTURE WAS DETERMINED USING TWO AND THREE DIMENSIONAL NMR EXPERIMENTS ON, RESPECTIVELY, UNLABELLED AND N-15 LABELLED TFF1 DIMER PROTEIN. MANY ASSIGN WERE MADE BY MAKING USE IF THE BETTER RESOLVED MONOMER SPECTRA (POLSHAKOV ET AL. 1995,1997) |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 0.01 / pH: 5.9 / Pressure: 1 atm / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | ||||||||||||
| NMR ensemble | Conformer selection criteria: TOTAL NOE CONSTRAINT VIOLATIONS < 1.0 A Conformers calculated total number: 11 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller








 PDBj
PDBj

 HSQC
HSQC