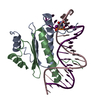
Entry Database : PDB / ID : 1hbxTitle Ternary Complex of SAP-1 and SRF with specific SRE DNA 5'-D(*CP*AP*CP*AP*CP*CP*GP*GP*AP*AP*GP*TP*CP* CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*T)-3'5'-D(*GP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP* GP*GP*AP*CP*TP*TP*CP*CP*GP*GP*TP*G)-3'ETS-DOMAIN PROTEIN ELK-4 SERUM RESPONSE FACTOR Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / / Resolution : 3.15 Å Authors Hassler, M. / Richmond, T.J. Journal : Embo J. / Year : 2001Title : The B-Box Dominates Sap-1/Srf Interactions in the Structure of the Ternary ComplexAuthors : Hassler, M. / Richmond, T.J. History Deposition Apr 20, 2001 Deposition site / Processing site Revision 1.0 Jun 27, 2001 Provider / Type Revision 1.1 May 7, 2011 Group Revision 1.2 Jul 13, 2011 Group Revision 1.3 Dec 13, 2023 Group Data collection / Database references ... Data collection / Database references / Other / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.15 Å
MOLECULAR REPLACEMENT / Resolution: 3.15 Å  Authors
Authors Citation
Citation Journal: Embo J. / Year: 2001
Journal: Embo J. / Year: 2001 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1hbx.cif.gz
1hbx.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1hbx.ent.gz
pdb1hbx.ent.gz PDB format
PDB format 1hbx.json.gz
1hbx.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hb/1hbx
https://data.pdbj.org/pub/pdb/validation_reports/hb/1hbx ftp://data.pdbj.org/pub/pdb/validation_reports/hb/1hbx
ftp://data.pdbj.org/pub/pdb/validation_reports/hb/1hbx Links
Links Assembly
Assembly


 Components
Components HOMO SAPIENS (human) / Gene: SRF (132-223) / Plasmid: PET3A / Gene (production host): SRF CORE (132-223) / Production host:
HOMO SAPIENS (human) / Gene: SRF (132-223) / Plasmid: PET3A / Gene (production host): SRF CORE (132-223) / Production host: 
 HOMO SAPIENS (human)
HOMO SAPIENS (human) HOMO SAPIENS (human) / Gene: SAP-1 (1-156) / Plasmid: PET3A / Gene (production host): SAP-1 (1-156) / Production host:
HOMO SAPIENS (human) / Gene: SAP-1 (1-156) / Plasmid: PET3A / Gene (production host): SAP-1 (1-156) / Production host: 
 HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID14-2 / Wavelength: 0.9326
/ Beamline: ID14-2 / Wavelength: 0.9326  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller















 PDBj
PDBj














































