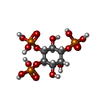
Entry Database : PDB / ID : 1h0aTitle Epsin ENTH bound to Ins(1,4,5)P3 EPSIN Keywords / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species RATTUS NORVEGICUS (Norway rat)Method / / / Resolution : 1.7 Å Authors Ford, M.G.J. / McMahon, H.T. / Evans, P.R. Journal : Nature / Year : 2002Title : Curvature of Clathrin-Coated Pits Driven by EpsinAuthors : Ford, M.G.J. / Mills, I. / Peter, B. / Vallis, Y. / Praefcke, G. / Evans, P.R. / Mcmahon, H.T. History Deposition Jun 12, 2002 Deposition site / Processing site Revision 1.0 Oct 3, 2002 Provider / Type Revision 1.1 May 8, 2011 Group Revision 1.2 Jul 13, 2011 Group Revision 1.3 Dec 13, 2023 Group Data collection / Database references ... Data collection / Database references / Other / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å
MOLECULAR REPLACEMENT / Resolution: 1.7 Å  Authors
Authors Citation
Citation Journal: Nature / Year: 2002
Journal: Nature / Year: 2002 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1h0a.cif.gz
1h0a.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1h0a.ent.gz
pdb1h0a.ent.gz PDB format
PDB format 1h0a.json.gz
1h0a.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/h0/1h0a
https://data.pdbj.org/pub/pdb/validation_reports/h0/1h0a ftp://data.pdbj.org/pub/pdb/validation_reports/h0/1h0a
ftp://data.pdbj.org/pub/pdb/validation_reports/h0/1h0a
 Links
Links Assembly
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID29 / Wavelength: 0.98
/ Beamline: ID29 / Wavelength: 0.98  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj