+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fyn | ||||||
|---|---|---|---|---|---|---|---|
| Title | PHOSPHOTRANSFERASE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / PROTO-ONCOGENE / TYROSINE-PROTEIN KINASE / PHOSPHORYLATION / ATP-BINDING / MYRISTYLATION / SH3 DOMAIN / COMPLEX (PHOSPHOTRANSFERASE-PEPTIDE) | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of hydrogen peroxide biosynthetic process / response to singlet oxygen / Reelin signalling pathway / perinuclear endoplasmic reticulum / NTRK2 activates RAC1 / regulation of glutamate receptor signaling pathway / Activated NTRK2 signals through FYN / heart process / SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion / regulation of calcium ion import across plasma membrane ...negative regulation of hydrogen peroxide biosynthetic process / response to singlet oxygen / Reelin signalling pathway / perinuclear endoplasmic reticulum / NTRK2 activates RAC1 / regulation of glutamate receptor signaling pathway / Activated NTRK2 signals through FYN / heart process / SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion / regulation of calcium ion import across plasma membrane / Platelet Adhesion to exposed collagen / reelin-mediated signaling pathway / G protein-coupled glutamate receptor signaling pathway / CRMPs in Sema3A signaling / FLT3 signaling through SRC family kinases / activated T cell proliferation / type 5 metabotropic glutamate receptor binding / positive regulation of protein localization to membrane / CD4 receptor binding / feeding behavior / cellular response to L-glutamate / Nef and signal transduction / Co-stimulation by CD28 / EPH-Ephrin signaling / natural killer cell activation / Nephrin family interactions / negative regulation of dendritic spine maintenance / DCC mediated attractive signaling / dendritic spine maintenance / Ephrin signaling / CD28 dependent Vav1 pathway / cellular response to peptide hormone stimulus / growth factor receptor binding / Regulation of KIT signaling / tau-protein kinase activity / leukocyte migration / phospholipase activator activity / EPHA-mediated growth cone collapse / Co-inhibition by CTLA4 / dendrite morphogenesis / peptide hormone receptor binding / CD8 receptor binding / Dectin-2 family / stimulatory C-type lectin receptor signaling pathway / Fc-gamma receptor signaling pathway involved in phagocytosis / forebrain development / PECAM1 interactions / response to amyloid-beta / cellular response to glycine / FCGR activation / Sema3A PAK dependent Axon repulsion / EPH-ephrin mediated repulsion of cells / CD28 dependent PI3K/Akt signaling / glial cell projection / Role of LAT2/NTAL/LAB on calcium mobilization / positive regulation of protein targeting to membrane / vascular endothelial growth factor receptor signaling pathway / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / ephrin receptor signaling pathway / detection of mechanical stimulus involved in sensory perception of pain / cellular response to transforming growth factor beta stimulus / alpha-tubulin binding / postsynaptic density, intracellular component / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / T cell receptor binding / phosphatidylinositol 3-kinase binding / ephrin receptor binding / GPVI-mediated activation cascade / phospholipase binding / T cell costimulation / cellular response to platelet-derived growth factor stimulus / Signaling by ERBB2 / EPHB-mediated forward signaling / NCAM signaling for neurite out-growth / negative regulation of protein ubiquitination / CD209 (DC-SIGN) signaling / axon guidance / cell surface receptor protein tyrosine kinase signaling pathway / FCGR3A-mediated IL10 synthesis / peptidyl-tyrosine phosphorylation / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / negative regulation of angiogenesis / Cell surface interactions at the vascular wall / learning / non-membrane spanning protein tyrosine kinase activity / FCGR3A-mediated phagocytosis / actin filament / non-specific protein-tyrosine kinase / Regulation of signaling by CBL / negative regulation of inflammatory response to antigenic stimulus / protein catabolic process / Signaling by SCF-KIT / positive regulation of neuron projection development / G protein-coupled receptor binding / negative regulation of protein catabolic process / modulation of chemical synaptic transmission / positive regulation of protein localization to nucleus / VEGFA-VEGFR2 Pathway / tau protein binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.3 Å X-RAY DIFFRACTION / Resolution: 2.3 Å | ||||||
 Authors Authors | Musacchio, A. / Saraste, M. / Wilmanns, M. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1994 Journal: Nat.Struct.Biol. / Year: 1994Title: High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Authors: Musacchio, A. / Saraste, M. / Wilmanns, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fyn.cif.gz 1fyn.cif.gz | 26.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fyn.ent.gz pdb1fyn.ent.gz | 16.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fyn.json.gz 1fyn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fy/1fyn https://data.pdbj.org/pub/pdb/validation_reports/fy/1fyn ftp://data.pdbj.org/pub/pdb/validation_reports/fy/1fyn ftp://data.pdbj.org/pub/pdb/validation_reports/fy/1fyn | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 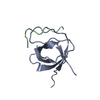
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6953.491 Da / Num. of mol.: 1 / Fragment: SH3 DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Strain: BL21 DE3 / Gene: FYN TYROSINE KINASE / Production host: Homo sapiens (human) / Strain: BL21 DE3 / Gene: FYN TYROSINE KINASE / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 1031.201 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: CHEMICALLY SYNTHESIZED |
| #3: Water | ChemComp-HOH / |
| Compound details | THIS ENTRY CONTAINS COORDINATES OF THE COMPLEX BETWEEN THE SH3 DOMAIN OF FYN TYROSINE KINASE AND A ...THIS ENTRY CONTAINS COORDINATE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.86 Å3/Da / Density % sol: 33.86 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 8.2 / Method: vapor diffusion, hanging dropDetails: protein solution is mixed with lyophilized 3BP-2 peptide at a 1:1 molar ratio and subsequently mixed at 1:1(v:v) to reservoir solution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 2.3 Å / % possible obs: 88 % / Rmerge(I) obs: 0.088 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor Rwork: 0.234 / Rfactor obs: 0.234 / Highest resolution: 2.3 Å Details: THE 3BP2 PEPTIDE LIES ON A CRYSTALLOGRAPHIC TWO-FOLD AXIS, I.E., IT IS DISORDERED. OCCUPANCY OF PEPTIDE ATOMS HAS BEEN SET TO 0.5. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2.3 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 8 Å / Num. reflection obs: 2436 / Rfactor obs: 0.171 / Rfactor Rwork: 0.171 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: x_bond_d / Dev ideal: 0.011 |
 Movie
Movie Controller
Controller















 PDBj
PDBj




















