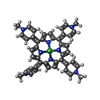
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1em0 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | COMPLEX OF D(CCTAGG) WITH TETRA-[N-METHYL-PYRIDYL] PORPHYRIN | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / PORPHYRIN / RUFFLING / DNA DISTORTION / GROOVE BINDING | Function / homology | TETRA[N-METHYL-PYRIDYL] PORPHYRIN-NICKEL / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 0.9 Å SYNCHROTRON / Resolution: 0.9 Å  Authors AuthorsNeidle, S. / Sanderson, M. / Bennett, M. / Krah, A. / Wien, F. / Garman, E. / McKenna, R. |  Citation Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2000 Journal: Proc.Natl.Acad.Sci.USA / Year: 2000Title: A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling. Authors: Bennett, M. / Krah, A. / Wien, F. / Garman, E. / McKenna, R. / Sanderson, M. / Neidle, S. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1em0.cif.gz 1em0.cif.gz | 46.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1em0.ent.gz pdb1em0.ent.gz | 33.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1em0.json.gz 1em0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1em0_validation.pdf.gz 1em0_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1em0_full_validation.pdf.gz 1em0_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  1em0_validation.xml.gz 1em0_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  1em0_validation.cif.gz 1em0_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/em/1em0 https://data.pdbj.org/pub/pdb/validation_reports/em/1em0 ftp://data.pdbj.org/pub/pdb/validation_reports/em/1em0 ftp://data.pdbj.org/pub/pdb/validation_reports/em/1em0 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 1888.113 Da / Num. of mol.: 4 / Source method: obtained synthetically #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.14 Å3/Da / Density % sol: 42.44 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 290 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1.25UM DNA, 1.25UM NI-PORPHYRIN, 50UM MGCL2, 5% MPD, 30MM SODIUM CACODYLATE, AGAINST 500UM RESERVOIR OF 55% MPD, pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 290K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||
| Detector |
| |||||||||||||||
| Radiation |
| |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Resolution: 0.86→30 Å / Num. obs: 45197 / % possible obs: 93 % / Observed criterion σ(F): 4 / Observed criterion σ(I): 2 / Redundancy: 8 % / Rmerge(I) obs: 0.021 | |||||||||||||||
| Reflection shell | Resolution: 0.9→1 Å / Redundancy: 8 % / Rmerge(I) obs: 0.124 / % possible all: 70.3 | |||||||||||||||
| Reflection | *PLUS Num. measured all: 336951 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 0.9→6 Å / Num. parameters: 3867 / Num. restraintsaints: 0 / Cross valid method: THROUGHOUT / σ(F): 4 / σ(I): 2 Stereochemistry target values: NO RESTRAINTS USED, THEREFORE THE RMSD DEVIATIONS ABOVE DO NOT HAVE ANY MEANING Details: FULL-MATRIX LEAST-SQUARES
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.9→6 Å
| |||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 0.9 Å / Lowest resolution: 6 Å / σ(F): 4 / % reflection Rfree: 5 % / Rfactor obs: 0.142 / Rfactor Rfree: 0.161 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller










 PDBj
PDBj