[English] 日本語
 Yorodumi
Yorodumi- PDB-1ss7: Compensating bends in a 16 base-pair DNA oligomer containing a T3... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ss7 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | ||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / B-DNA / double helix / Residual dipolar couplings | Function / homology | DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / Restrained molecular dynamics |  Authors AuthorsMcAteer, K. / Aceves-Gaona, A. / Michalczyk, R. / Buchko, G.W. / Isern, N.G. / Silks, L.A. / Miller, J.H. / Kennedy, M.A. |  Citation Citation Journal: Biopolymers / Year: 2004 Journal: Biopolymers / Year: 2004Title: Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature Authors: McAteer, K. / Aceves-Gaona, A. / Michalczyk, R. / Buchko, G.W. / Isern, N.G. / Silks, L.A. / Miller, J.H. / Kennedy, M.A. History |
|
- Structure visualization
Structure visualization
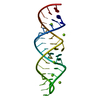
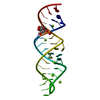
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ss7.cif.gz 1ss7.cif.gz | 302.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ss7.ent.gz pdb1ss7.ent.gz | 248.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ss7.json.gz 1ss7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ss/1ss7 https://data.pdbj.org/pub/pdb/validation_reports/ss/1ss7 ftp://data.pdbj.org/pub/pdb/validation_reports/ss/1ss7 ftp://data.pdbj.org/pub/pdb/validation_reports/ss/1ss7 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 4898.191 Da / Num. of mol.: 2 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: Heteronuclear dipolar couplings were measured in samples aligned with filamentous Pf1 bacteriophage |
- Sample preparation
Sample preparation
| Details | Contents: 1 mM DNA / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 400 mM NaCl, 40mM phosphate / pH: 7 / Pressure: ambient / Temperature: 308 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Restrained molecular dynamics / Software ordinal: 1 Details: Structures are based on a total of 316 NOE, 90 torsion angle, and 6 distance restraints represented each W-C base-pair. For the RDC structures an additional 88 one-bond C-H and 14 one-bond N- ...Details: Structures are based on a total of 316 NOE, 90 torsion angle, and 6 distance restraints represented each W-C base-pair. For the RDC structures an additional 88 one-bond C-H and 14 one-bond N-H residual dipolar coupling restraints were used | ||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 45 / Conformers submitted total number: 15 |
 Movie
Movie Controller
Controller











 PDBj
PDBj






































 HSQC
HSQC