[English] 日本語
 Yorodumi
Yorodumi- PDB-1ekz: NMR STRUCTURE OF THE COMPLEX BETWEEN THE THIRD DSRBD FROM DROSOPH... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ekz | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR STRUCTURE OF THE COMPLEX BETWEEN THE THIRD DSRBD FROM DROSOPHILA STAUFEN AND A RNA HAIRPIN | ||||||
 Components Components |
| ||||||
 Keywords Keywords | cell cycle/RNA / protein/RNA / protein dsRBD / Drosophila / RNA hairpin / cell cycle-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationgerm plasm / subsynaptic reticulum / pole plasm RNA localization / bicoid mRNA localization / regulation of oskar mRNA translation / pole plasm protein localization / posterior cell cortex / regulation of pole plasm oskar mRNA localization / asymmetric protein localization involved in cell fate determination / anterograde dendritic transport of messenger ribonucleoprotein complex ...germ plasm / subsynaptic reticulum / pole plasm RNA localization / bicoid mRNA localization / regulation of oskar mRNA translation / pole plasm protein localization / posterior cell cortex / regulation of pole plasm oskar mRNA localization / asymmetric protein localization involved in cell fate determination / anterograde dendritic transport of messenger ribonucleoprotein complex / neuroblast fate determination / positive regulation of cytoplasmic mRNA processing body assembly / pole plasm assembly / positive regulation of synaptic assembly at neuromuscular junction / neuronal ribonucleoprotein granule / RNA localization / basal cortex / anterior/posterior axis specification, embryo / apical cortex / intracellular mRNA localization / protein localization to synapse / oogenesis / lncRNA binding / germ cell development / long-term memory / dendrite cytoplasm / basal plasma membrane / mRNA 3'-UTR binding / P-body / cytoplasmic ribonucleoprotein granule / cytoplasmic stress granule / apical part of cell / double-stranded RNA binding / cell cortex / neuron projection / apical plasma membrane / neuronal cell body / mRNA binding / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Ramos, A. / Grunert, S. / Bycroft, M. / St Johnston, D. / Varani, G. | ||||||
 Citation Citation |  Journal: EMBO J. / Year: 2000 Journal: EMBO J. / Year: 2000Title: RNA recognition by a Staufen double-stranded RNA-binding domain. Authors: Ramos, A. / Grunert, S. / Adams, J. / Micklem, D.R. / Proctor, M.R. / Freund, S. / Bycroft, M. / St Johnston, D. / Varani, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
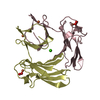
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ekz.cif.gz 1ekz.cif.gz | 1.5 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ekz.ent.gz pdb1ekz.ent.gz | 1.3 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ekz.json.gz 1ekz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ekz_validation.pdf.gz 1ekz_validation.pdf.gz | 384 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ekz_full_validation.pdf.gz 1ekz_full_validation.pdf.gz | 904 KB | Display | |
| Data in XML |  1ekz_validation.xml.gz 1ekz_validation.xml.gz | 86.3 KB | Display | |
| Data in CIF |  1ekz_validation.cif.gz 1ekz_validation.cif.gz | 131.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ek/1ekz https://data.pdbj.org/pub/pdb/validation_reports/ek/1ekz ftp://data.pdbj.org/pub/pdb/validation_reports/ek/1ekz ftp://data.pdbj.org/pub/pdb/validation_reports/ek/1ekz | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 9612.737 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: Protein | Mass: 8463.958 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations, structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 36 |
|---|
 Movie
Movie Controller
Controller









 PDBj
PDBj





























