[English] 日本語
 Yorodumi
Yorodumi- PDB-1eht: THEOPHYLLINE-BINDING RNA IN COMPLEX WITH THEOPHYLLINE, NMR, 10 ST... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1eht | ||||||
|---|---|---|---|---|---|---|---|
| Title | THEOPHYLLINE-BINDING RNA IN COMPLEX WITH THEOPHYLLINE, NMR, 10 STRUCTURES | ||||||
 Components Components | THEOPHYLLINE-BINDING RNA | ||||||
 Keywords Keywords | RNA / RIBONUCLEIC ACID | ||||||
| Function / homology | THEOPHYLLINE / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Zimmermann, G.R. / Pardi, A. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Authors: Zimmermann, G.R. / Jenison, R.D. / Wick, C.L. / Simorre, J.P. / Pardi, A. #1:  Journal: Science / Year: 1994 Journal: Science / Year: 1994Title: High-Resolution Molecular Discrimination by RNA Authors: Jenison, R.D. / Gill, S.C. / Pardi, A. / Polisky, B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1eht.cif.gz 1eht.cif.gz | 216 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1eht.ent.gz pdb1eht.ent.gz | 176.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1eht.json.gz 1eht.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eh/1eht https://data.pdbj.org/pub/pdb/validation_reports/eh/1eht ftp://data.pdbj.org/pub/pdb/validation_reports/eh/1eht ftp://data.pdbj.org/pub/pdb/validation_reports/eh/1eht | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
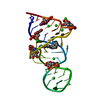
| Deposited unit | 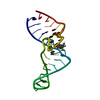
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 10638.409 Da / Num. of mol.: 1 / Fragment: BINDING FRAGMENT, RESIDUES 1 - 33 / Source method: obtained synthetically Details: CONSENSUS SEQUENCE OF THE THEOPHYLLINE BINDING RNA APTAMER FLANKED BY A FOUR BASE-PAIR STEM, A THREE BASE-PAIR STEM, AND CAPPED BY A GAAA TETRALOOP |
|---|---|
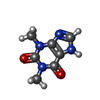
| #2: Chemical | ChemComp-TEP / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Sample conditions | pH: 6.8 / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 500 MHz |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST TOTAL ENERGY / Conformers calculated total number: 50 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller








 PDBj
PDBj






























 X-PLOR
X-PLOR