+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ea4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | GENE REGULATION/DNA / TRANSCRIPTIONAL REPRESSOR / DNA-BINDING PROTEIN / PLASMID / PROTEIN-DNA COMPLEX / GENE REGULATION-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationplasmid maintenance / protein-DNA complex / sequence-specific DNA binding / regulation of DNA-templated transcription Similarity search - Function | ||||||
| Biological species |  STREPTOCOCCUS AGALACTIAE (bacteria) STREPTOCOCCUS AGALACTIAE (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.95 Å MOLECULAR REPLACEMENT / Resolution: 2.95 Å | ||||||
 Authors Authors | Gomis-Rueth, F.X. / Costa, M. / Sola, M. / Acebo, P. / Eritja, R. / Espinosa, M. / Solar, G.D. / Coll, M. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides Authors: Costa, M. / Sola, M. / Del, G. / Eritja, R. / Hernaindez-Arriaga, A.M. / Espinosa, M. / Gomis-Rueth, F.X. / Coll, M. #1:  Journal: Embo J. / Year: 1998 Journal: Embo J. / Year: 1998Title: The Structure of Plasmid-Encoded Transcriptional Repressor Copg Unliganded and Bound to its Operator Authors: Gomis-Rueth, F.X. / Sola, M. / Acebo, P. / Parraga, A. / Guasch, A. / Eritja, R. / Gonzalez, A. / Espinosa, M. / Solar, G.D. / Coll, M. #2: Journal: FEBS Lett. / Year: 1998 Title: Overexpression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of the Pmv158-Encoded Plasmid Transcriptional Repressor Protein Copg Authors: Gomis-Rueth, F.X. / Sola, M. / Perez-Luque, R. / Acebo, P. / Alda, M.T. / Gonzalez, A. / Espinosa, M. / Solar, G.D. / Coll, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ea4.cif.gz 1ea4.cif.gz | 160.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ea4.ent.gz pdb1ea4.ent.gz | 124.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ea4.json.gz 1ea4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ea/1ea4 https://data.pdbj.org/pub/pdb/validation_reports/ea/1ea4 ftp://data.pdbj.org/pub/pdb/validation_reports/ea/1ea4 ftp://data.pdbj.org/pub/pdb/validation_reports/ea/1ea4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1b01S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 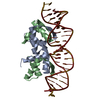
| ||||||||
| Unit cell |
| ||||||||
| Details | FUNCTIONAL TETRAMERS (EACH ONE CONTACTING A 22BP DSDNA)ARE DEFG, HJKL, AND ABA'B' (A' AND B' ARE SYMMETRYEQUIVALENT MOLECULES).TETRAMER DEFG CONTACTS DSDNA WX, HJKL PAIRS UV, ANDABA'B' INTERACTS WITH YZ(DOUBLE OCCUPANCY DUE TO CRYSTALLOGRAPHIC TWOFOLD AXIS)THE BIOMOLECULE 1 IS THE SUPERHELICAL STRUCTURE AND THETETRAMERS CAN BE GENERATED USING THE MATRICES GIVENFOR BIOMOLECULES 2 , 3 AND 4 |
- Components
Components
| #1: Protein/peptide | Mass: 5124.092 Da / Num. of mol.: 10 / Fragment: DNA-BINDING PROTEIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)  STREPTOCOCCUS AGALACTIAE (bacteria) / Cellular location: PLASMID PMV158 / Plasmid: PMV158 / Production host: STREPTOCOCCUS AGALACTIAE (bacteria) / Cellular location: PLASMID PMV158 / Plasmid: PMV158 / Production host:  #2: DNA chain | Mass: 6680.344 Da / Num. of mol.: 3 / Fragment: 22BP SSDNA - FIRST STRAND / Source method: obtained synthetically #3: DNA chain | Mass: 6822.412 Da / Num. of mol.: 3 / Fragment: 22BP SSDNA - SECOND STRAND / Source method: obtained synthetically #4: Water | ChemComp-HOH / | Compound details | REGULATES THE PLASMID COPY NUMBER BY BINDING TO THE REPAB PROMOTER THUS CONTROLING THE SYNTHESIS OF ...REGULATES THE PLASMID COPY NUMBER BY BINDING TO THE REPAB PROMOTER THUS CONTROLING | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 44.94 % Description: ONE COPG DIMER/ 9BP DSDNA MODEL WAS USED AS SEARCHING MODEL. | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.6 / Details: MPD, NACL, NAACO, pH 4.60 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1.0527 / Beamline: 5.2R / Wavelength: 1.0527 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0527 Å / Relative weight: 1 |
| Reflection | Resolution: 2.92→43.85 Å / Num. obs: 17592 / % possible obs: 93.6 % / Redundancy: 5.9 % / Biso Wilson estimate: 80.8 Å2 / Rmerge(I) obs: 0.106 / Net I/σ(I): 5.9 |
| Reflection | *PLUS Num. measured all: 103432 |
| Reflection shell | *PLUS Highest resolution: 2.92 Å / Lowest resolution: 3.07 Å / % possible obs: 58.6 % / Rmerge(I) obs: 0.471 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1B01 Resolution: 2.95→40 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: NOE RESTRAINTS FOR WATSON & CRICK BASE PAIRING. THE COMPLEX SET UP FOR CRYSTALLIZATION WAS MADE UP BY A COPG DIMER-OF-HOMODIMERS AND A 22-BP DSDNA. THERE ARE 2,5 OF THOSE COMPLEXES IN THE ...Details: NOE RESTRAINTS FOR WATSON & CRICK BASE PAIRING. THE COMPLEX SET UP FOR CRYSTALLIZATION WAS MADE UP BY A COPG DIMER-OF-HOMODIMERS AND A 22-BP DSDNA. THERE ARE 2,5 OF THOSE COMPLEXES IN THE ASYMMETRIC UNIT, DEFG+WX (PROTEIN + DNA), HJKL+UV, AND ABA'B'+YZ. THE LATTER REPRESENTS THE "HALF" COMPLEX. THE OTHER HALF IS CREATED BY A CRYSTALLOGRAPHIC TWOFOLD (RENDERING A' AND B'). THE DNA PART HAS BEEN MODELLED WITH THE TWO OBSERVED ORIENTATIONS, EACH WITH OCCUPANCY 0.5. THERE ARE NCS RESTRAINTS, BUT SO MANY THAT THE MATRICES AND TRANSLATIONS HAVE NOT BEEN INCLUDED IN THIS ENTRY. ESSENTIALLY, ALL PROTEIN CHAINS AND ALL DNA STRANDS HAVE BEEN SUBJECTED TO RESTRAINTS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 66.3 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.95→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: RESTRAINTS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.9/1.0 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.23 / Rfactor Rwork: 0.23 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller










 PDBj
PDBj







































